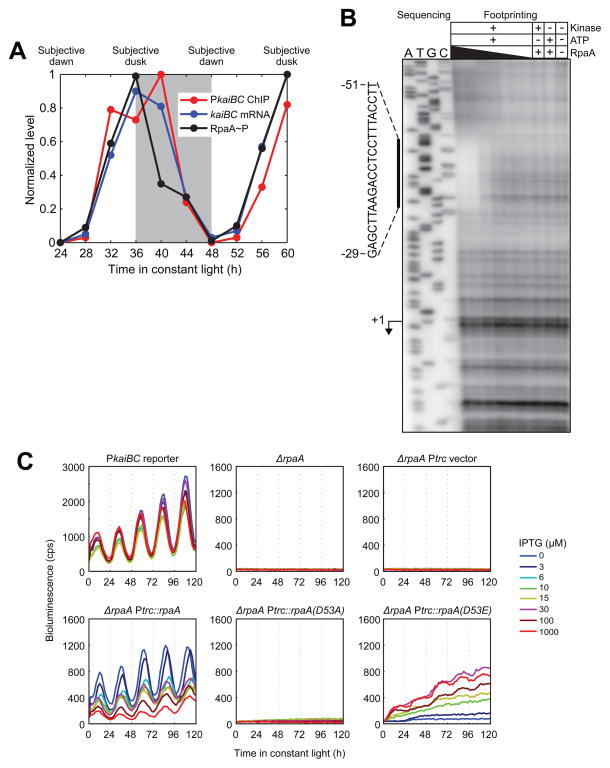
Figure 3. RpaA binds to the kaiBC promoter in vivo and in vitro and promoteskaiBC expression in a phosphorylation-dependent manner.
A. Correlation between RpaA phosphorylation, RpaA enrichment at the kaiBC promoter, and abundance of the kaiBC transcript. Subjective night is shaded. RpaA phosphorylation was measured by Phos-tag Western blot (see Figure S3B for gel image), association with PkaiBC by ChIP-qPCR, and kaiBC expression by RT-qPCR. Note that while the apex of the ChIP-qPCR enrichment is at 40 h (four hours after subjective dusk) in this experiment, the precise phase of RpaA (as well as that of global gene expression oscillations) varies between experiments, with RpaA binding typically peaking at or a few hours before subjective dusk.
B. In vitro DNase I footprinting of RpaA on the kaiBC promoter as a function of recombinant RpaA phosphorylation and concentration. Sanger sequencing reactions used to identify the location of the footprint are shown on the left; footprinting reactions are shown on the right. The region protected from digestion by RpaA~P is indicated by the vertical bar. The kaiBC transcription start site (Kutsuna et al., 2005) is indicated with an arrow. RpaA pre-treatment and concentration are indicated above each footprinting lane. RpaA was at least 50% phosphorylated in the presence of both CikA and ATP but was unphosphorylated otherwise (Figure S3D). RpaA was added to a final concentration of 6.0, 3.0, 0.6, 0.3, 0.06, or 0.03 μM as indicated by the thickness of the wedge.
C. Activity of the kaiBC promoter was assayed using a PkaiBC::luxAB luciferase reporter in various genetic backgrounds: wild-type, ΔrpaA, and ΔrpaA expressing wild-type RpaA, unphosphorylatable RpaA (D53A), or an RpaA phosphomimetic (D53E) from the IPTG-inducible Ptrc promoter. IPTG was added at the indicated concentration prior to entrainment with two 12-h dark pulses.
See also Figure S3.