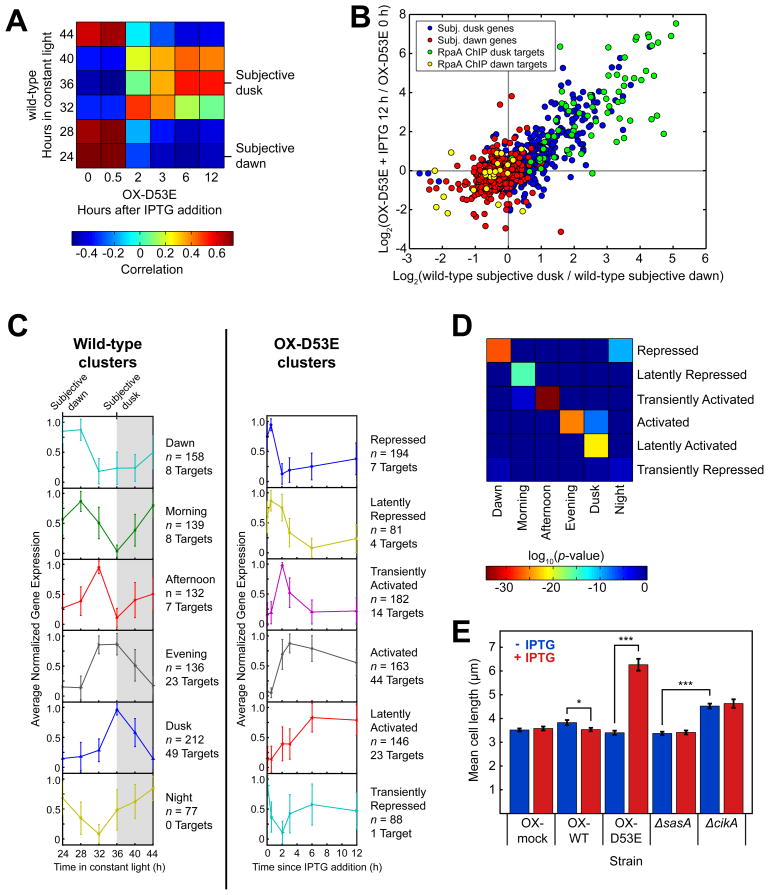
Figure 6. RpaA orchestrates global circadian gene expression and controls the cell division gate.
A. Correlation between the expression of circadian genes (n = 856) in the wild-type strain over the course of one day and the expression of those genes in the ΔrpaA ΔkaiBC Ptrc::rpaA(D53E) (OX-D53E) strain before (t = 0 h) and after induction with IPTG. Gene expression was measured by RNA-Seq.
B. Correlation between the change in expression of circadian genes (n = 856) caused by induction of RpaA(D53E) in the OX-D53E strain (y-axis) with the change in expression between subjective dusk and dawn in the wild-type strain (x-axis). RpaA ChIP target genes are highlighted (n = 95; 71 subjective dusk and 24 subjective dawn). Gene expression was measured by RNA-Seq.
C. K-means identification of gene expression clusters in the wild-type and OX-D53E strains. Gene expression was measured by RNA-Seq. With K = 6, wild-type circadian genes (n = 856) were separated into six clusters with distinct expression phases (left), consistent with previous microarray observations (Vijayan et al., 2009). Timecourses of the same set of genes in the OX-D53E strain were also clustered using K = 6 (right). The traces show the average normalized timecourse of genes within each cluster; error bars show standard deviation. The numbers of all genes (n) and RpaA ChIP target genes in each cluster are indicated. Non-coding RNAs were not included in this analysis.
D. Mapping between clusters in the wild-type (x-axis) and OX-D53E (y-axis) strains. Each element of the heatmap shows the log10 of the statistical significance (Fisher’s exact test) of the overlap between the corresponding clusters on each axis.
E. Mean cell lengths in ΔrpaA ΔkaiBC strains containing a Ptrc promoter driving expression of wild-type RpaA (OX-WT), RpaA(D53E) (OX-D53E), or an empty multi-cloning site (OX-mock) grown in the presence (red) or absence (blue) of the inducer IPTG (100 μM). At least 80 cells were analyzed for each strain. Error bars show standard error of the mean (SEM). *, p < 0.05; ***, p < 10−13 (one-way ANOVA).