Abstract
To investigate the pattern of allelic distribution in enzyme polymorphism, with special reference to the relationship between the mean (H) and the variance (VH) of heterozygosity, we used the model of effectively neutral mutations involving multiple alleles in which selective disadvantage of mutant alleles follows a Γ distribution. A simulation method was developed that enables us to study efficiently the process of random drift in a multiallelic genetic system and that saves a great deal of computer time. It is an improved version of the pseudosampling-variable (PSV) method [Kimura, M. (1980) Proc. Natl. Acad. Sci. USA 77, 522-526] previously used to simulate random drift in a diallelic system. This method will be useful for simulating many models of population genetics that involve behavior of multiple alleles in a finite population. By using this method, it was shown that, as compared with the model of strictly neutral mutations, the present model gives the reduction of both H and VH and an excess of rare variant alleles. The results were discussed in the light of recent observations on protein polymorphism with special reference to the functional constraint of proteins involved.
Keywords: population genetics, neutral theory of molecular evolution, polyallelic random drift
Full text
PDF

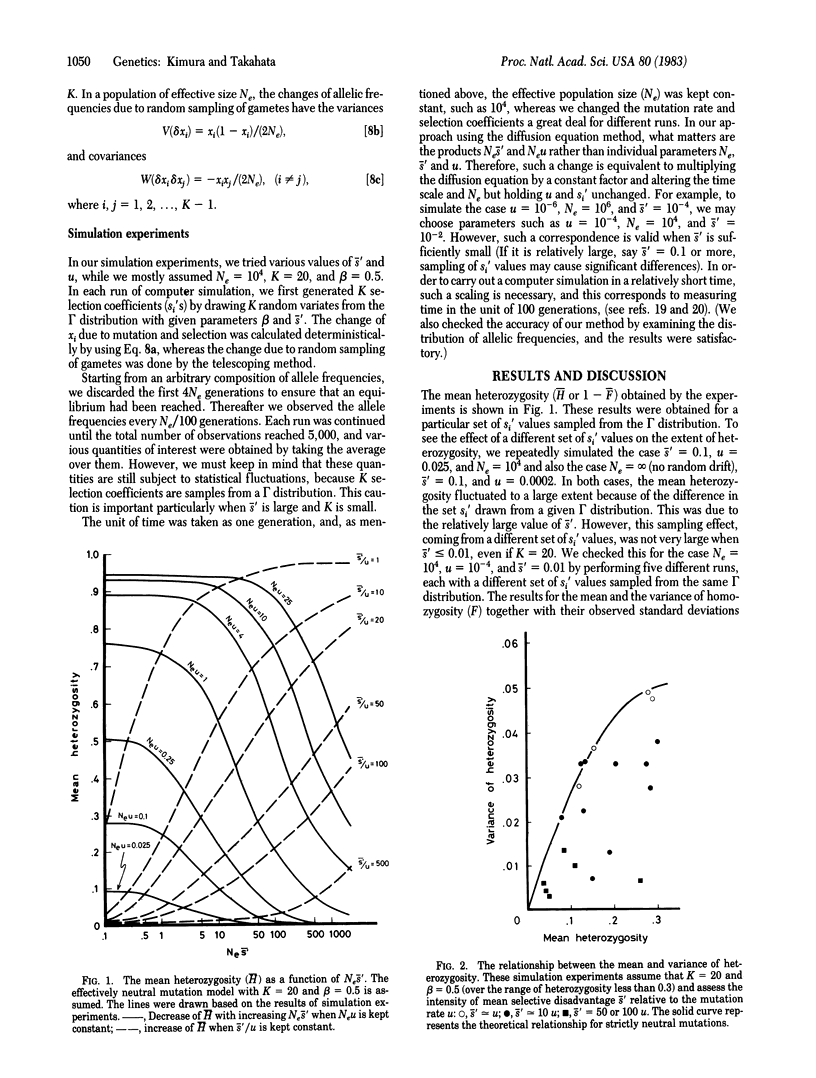
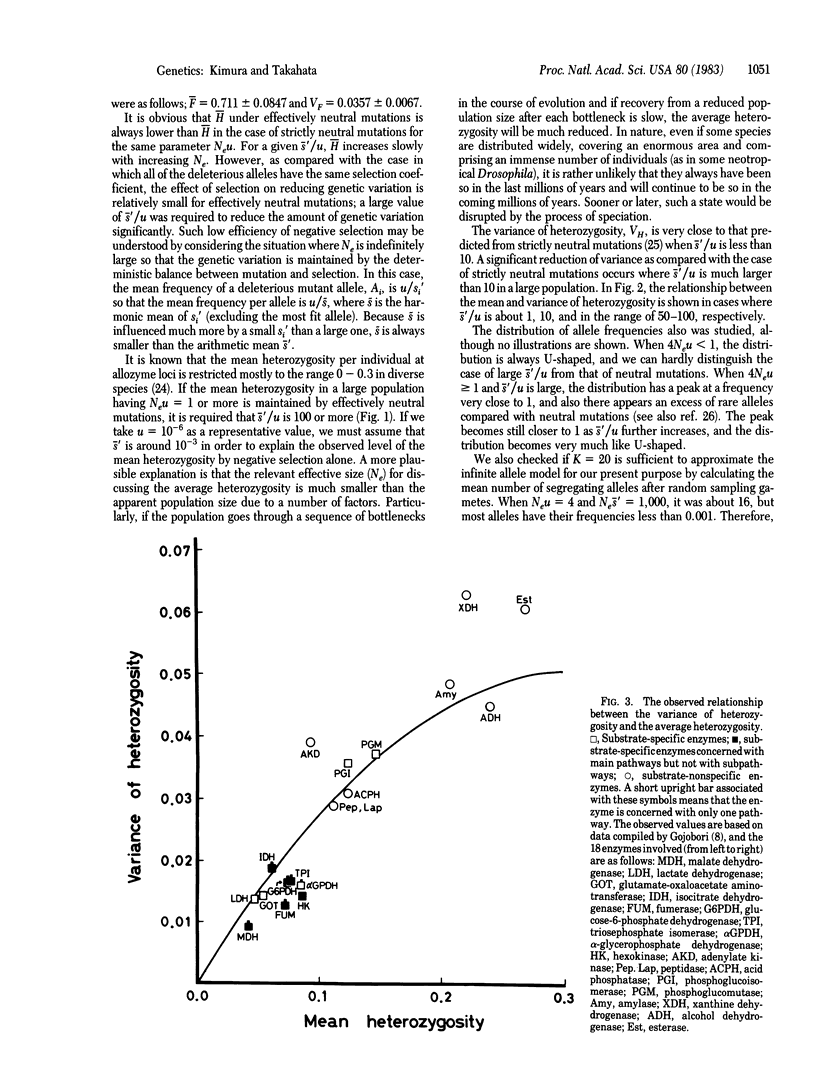

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fuerst P. A., Chakraborty R., Nei M. Statistical studies on protein polymorphism in natural populations. I. Distribution of single locus heterozygosity. Genetics. 1977 Jun;86(2 Pt 1):455–483. [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., Langley C. H. A general model to account for enzyme variation in natural populations. Genetics. 1974 Apr;76(4):837–848. doi: 10.1093/genetics/76.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIMURA M., CROW J. F. THE NUMBER OF ALLELES THAT CAN BE MAINTAINED IN A FINITE POPULATION. Genetics. 1964 Apr;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Average time until fixation of a mutant allele in a finite population under continued mutation pressure: Studies by analytical, numerical, and pseudo-sampling methods. Proc Natl Acad Sci U S A. 1980 Jan;77(1):522–526. doi: 10.1073/pnas.77.1.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet Res. 1968 Jun;11(3):247–269. doi: 10.1017/s0016672300011459. [DOI] [PubMed] [Google Scholar]
- Kimura M. Model of effectively neutral mutations in which selective constraint is incorporated. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3440–3444. doi: 10.1073/pnas.76.7.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ohta T. Protein polymorphism as a phase of molecular evolution. Nature. 1971 Feb 12;229(5285):467–469. doi: 10.1038/229467a0. [DOI] [PubMed] [Google Scholar]
- Li W. H. Maintenance of genetic variability under mutation and selection pressures in a finite population. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2509–2513. doi: 10.1073/pnas.74.6.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Nei M. Genetic variability maintained by mutation and overdominant selection in finite populations. Genetics. 1981 Jun;98(2):441–459. doi: 10.1093/genetics/98.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Fuerst P. A., Chakraborty R. Testing the neutral mutation hypothesis by distribution of single locus heterozygosity. Nature. 1976 Aug 5;262(5568):491–493. doi: 10.1038/262491a0. [DOI] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973 Nov 9;246(5428):96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ota T. Mutational pressure as the main cause of molecular evolution and polymorphism. Nature. 1974 Nov 29;252(5482):351–354. doi: 10.1038/252351a0. [DOI] [PubMed] [Google Scholar]
- Ota T. Statistical analyses of Drosophila and human protein polymorphisms. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3194–3196. doi: 10.1073/pnas.72.8.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. G. An approximate method of sampling a multinomial population. Biometrics. 1973 Dec;29(4):814–821. [PubMed] [Google Scholar]
- Stewart F. M. Variability in the amount of heterozygosity maintained by neutral mutations. Theor Popul Biol. 1976 Apr;9(2):188–201. doi: 10.1016/0040-5809(76)90044-7. [DOI] [PubMed] [Google Scholar]
- Takahata N. Genetic Variability and Rate of Gene Substitution in a Finite Population under Mutation and Fluctuating Selection. Genetics. 1981 Jun;98(2):427–440. doi: 10.1093/genetics/98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A. Heterosis or neutrality? Genetics. 1977 Apr;85(4):789–814. doi: 10.1093/genetics/85.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]


