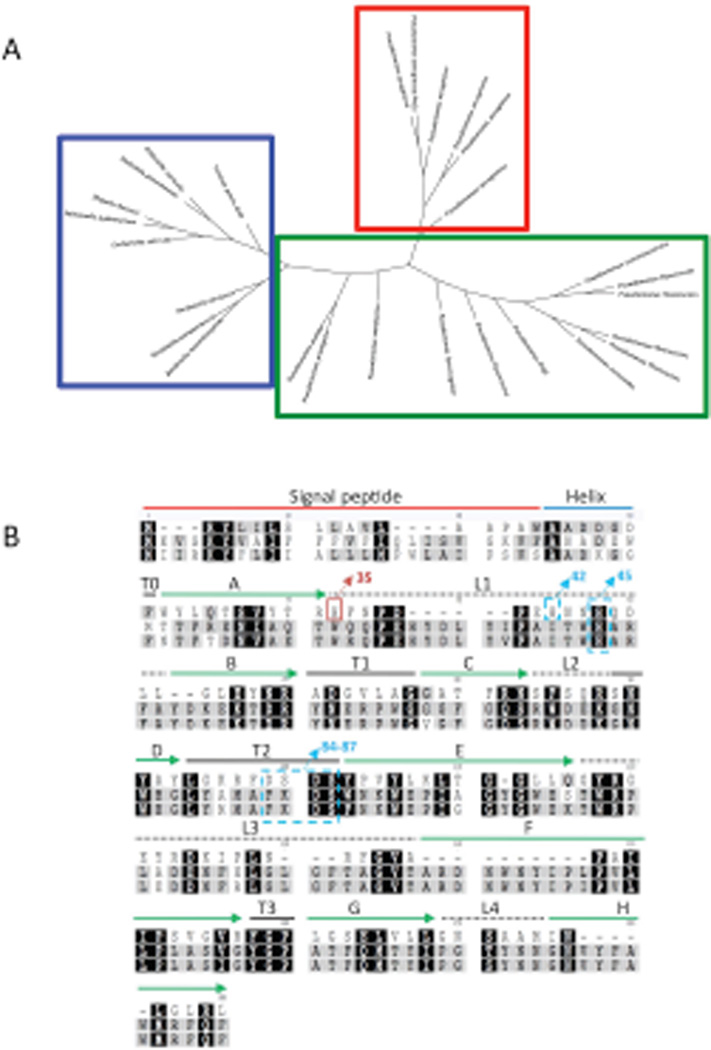
Fig. 4.
Tree diagram of the palmitoyltransferase enzymes of gram-negative bacteria and Pseudomonas species generated by using Geneious. (A) Clustering patterns show PA PagP (red box) is distinct and separate from other non-aeruginosa Pseudomonas PagP (green box), as well as other known PagP enzymes (blue box). (B) Alignment of PagP from three different pathogenic Gram-negative bacterial genera PA, E. coli and Salmonella typhimurium using ClustalW (www.geneious.com). Absolutely conserved residues are marked with black background. Residues indicated in grey denote weakly conserved residues, white indicates non-conserved residues. PA PagP domains are predicted based on the E. coli PagP sequence of the mature protein. Residues from 1-18AA represents the signal sequence, the predicted α-helix extends from 19AA to 37AA. The putative catalytic residues (H-42 and 45/D-84 and 86/S- 85 and 87) are indicated in blue dashed boxes. His35 in the red box indicates the catalytically necessary residue. Extracellular loops are indicated by dashed lines, solid green arrows indicate β-sheet.