Abstract
The mammalian high mobility group protein HMGA2 contains three DNA binding motifs associated with many physiological functions including oncogenesis, obesity, stem cell youth, human height, and human intelligence. In the present paper, trapped ion mobility spectrometry – mass spectrometry (TIMS-MS) has been utilized to study the conformational dynamics of the third DNA binding motif using the “AT hook” decapeptide unit (Lys1-Arg2-Prol3-Arg4-Gly5-Arg6-Prol7-Arg8-Lys9-Trp10, ATHP) as a function of the solvent state. Solvent state distributions were preserved during electrospray ion formation and multiple IMS bands were identified for the [M+2H]+2 and for the [M+3H]+3 charge states. Conformational isomer inter-conversion rates were measured as a function of the trapping time for the [M+2H]+2 and [M+3H]+3 charge states. Candidate structures were proposed for all IMS bands observed. Protonation site, proline residue conformation, and side chain orientations were identified as the main motifs governing the conformational inter-conversion processes. Conformational dynamics from the solvent state distribution to the gas-phase “de-solvated” state distribution demonstrated that ATHP is “structured”, and relative abundances are associated to the relative stability between the proposed conformers. The most stable ATHP [M+2H]+2 conformation at the “de-solvated” state corresponds to the AT-hook motif observed in AT-rich DNA regions.
Keywords: ion mobility spectrometry, peptide conformational dynamics
The mammalian high mobility group protein (HMGA2) is a multi-function nuclear transcription factor directly linked to oncogenesis1,2, obesity3,4 as well as human height5,6, stem cell youth7, and human intelligence8. HMGA2 is a small DNA-binding protein carrying three “AT hook” DNA binding motifs that specifically recognize the minor groove of AT-rich DNA sequences9. These DNA-binding motifs contain a consensus PRGRP sequence, flanked on each side by one of the positively charged amino acids, i.e., arginine or lysine. In the absence of AT-rich DNA, the “AT hook” DNA-binding motif is commonly assigned as “unstructured”.10,11 However, upon binding to the minor groove of five AT base pairs, it adopts a defined conformation.11,12 This disorder-to-order structure transition in HMGA2 accounts for a variety of nuclear activities, such as transcription, recombination, and DNA replication.8,9,12,13,14
With the advent of soft ionization techniques (e.g., electrospray ionization, ESI14), traditional structural analysis of biomolecules (e.g., circular dichroism, CD, and nuclear magnetic resonance spectroscopy, NMR) can be highly complemented with measurements of ion-neutral collision cross sections (CCSs) using ion mobility spectrometry (IMS).15–20 In particular, it has been shown that evaporative cooling of the solvent leads to a freezing of multiple conformations,20,21 which can be further experimentally analyzed and compared with theoretical calculations of candidate structures.21–23 With the recent introduction of trapped ion mobility spectrometry (TIMS),24,25 the latter studies can be extended as a function of the trapping time inside the IMS cell, thus permitting isomerization kinetic measurements. In the current study, it is reported for the first time the isomerization kinetics of a truncated form of HMGA2 containing the third DNA binding motif: a decapeptide consisting of Lys1-Arg2-Prol3-Arg4-Gly5-Arg6-Prol7-Arg8-Lys9-Trp10 (ATHP). The kinetic studies demonstrate that AT hook peptides can adopt multiple conformations in solution that vary according to charge state.
EXPERIMENTAL SECTION
The “AT hook” decapeptide unit (Lys1-Arg2-Prol3-Arg4-Gly5-Arg6-Prol7-Arg8-Lys9-Trp10, ATHP) was purchased from Advanced ChemTech Inc. (Louisville, KY) and used as received. Details on the TIMS-MS can be found elsewhere.24,25 Briefly, a TIMS analyzer was coupled to a maXis Impact Q-UHR-ToF (Bruker Daltonics Inc., Billerica, MA). Data acquisition was controlled using in-house software, written in National Instruments Lab VIEW, and synchronized with the maXis Impact acquisition program. Nitrogen was used as the bath gas at ca. 300K. All samples were prepared at 1µM concentration using HPLC grade solvents from Thermo Fisher Scientific Inc. (Waltham, MA). Mobility calibration and the number of isomer bands were determined using tune mix calibration standard (G2421A, Agilent Technologies, Santa Clara, CA) that comprises sphere-like structures that can only adopt one conformation (e.g., m/z = 322 Ko= 1.376 cm2V−1 s−1, m/z = 622 Ko= 1.013 cm2V−1 s−1, and m/z = 922 Ko= 0.835 cm2V−1 s−1).26
Mobility,K, of an ion in the TIMS cell can be described by:
| (1) |
where vg, E, Velution and Vbase are the velocity of the gas, applied electric field, elution and base voltages, respectively. The constant, A, is determined using standards of known mobilities under the same gas velocity conditions. The elution voltage, Velution, can be calculated from the elution time:
| (2) |
and
| (3) |
where Vo is the initial potential at the entrance to the TIMS analyzer, r is the rate at which the potential is ramped, Telution is the time at which the ion elutes, Tramp is the total ramp time, Ttotal is the total time for a single TIMS experiment, Ttrap is the time before the mobility analysis (i.e. to inject ions into the TIMS trap), and TOF is the time between elution of the ion and detection of the ion at the TOF detector. Mobility values (K) were correlated with CCS (Ω) using the equation:
| (4) |
where z is the charge of the ion, kB is the Boltzmann constant, N* is the number density and mI and mb refer to the masses of the molecular ion and bath gas molecule, respectively.
Molecular Dynamic Simulations
Conformational motifs were theoretically investigated using YASARA software (http://www.yasara.org). Briefly, simulations were performed using molecular mechanics (AMBER 03 force field) in a NVT thermostat that contains the molecular ion of interest interacting with nitrogen molecules. Typical simulation time was ~500 picoseconds and multiple charge state and protonation sites (i.e., amine terminal, arginine and lysine) were considered. The influence of the charge location for the [M+2]+2 and [M+3H]+3 is shown in the supporting information.
RESULTS AND DISCUSSION
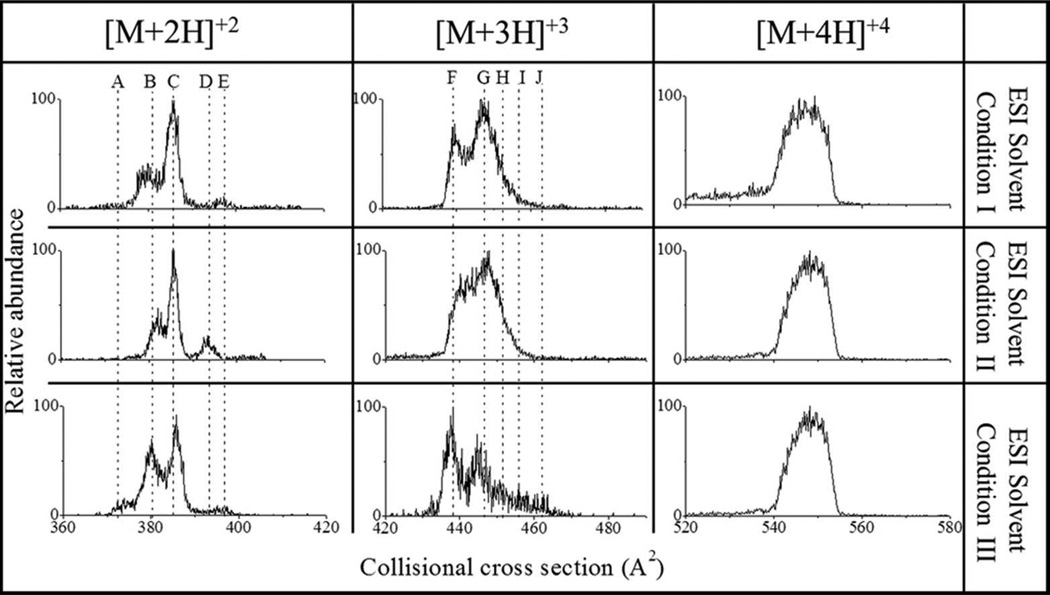
Changes in the IMS profile, particularly the relative abundance of IMS bands, of ATHP were observed as a function of the ESI solvent condition. Three ESI solution conditions are shown in Figure 1: water:methanol:acetonitrile (33:33:33), water:acetonitrile:acetic acid (49:49:2) and 10 mM ammonium acetate; from now on referred as ESI solvent conditions I, II and III, respectively. Other solutions, including water:methanol (0:100-100:0) were also compared; however, similar results as ESI solvent condition II were obtained, thus results are not presented here. Inspection of Figure 1 shows that various ATHP conformational isomers are likely preserved during the ESI process and reflect the initial solvent state distribution as in previous studies utilizing bradykinin peptides;23,28. Since CCS represents an “average” of the molecular ion surface that is accessible during collisions with the bath gas molecules, each IMS band corresponds to at least one conformational band (or family); that is, different conformations may yield the same CCS value. For example, ATHP [M+2H]+2 and [M+3H]+3 contain at least five conformation bands (labeled for each charge state), while ATHP [M+4H]+4 is dominated by a single broad band which is likely related to several conformations with very similar CCS values. When analyzed over time, changes in the IMS band distributions were observed for ATHP [M+2H]+2 and [M+3H]+3 but not for ATHP [M+4H]+4 regardless of the ESI solvent conditions. ESI solvent conditions I and II correspond to non-physiological solvent states, and for simplicity, further comparisons were conducted between ESI solvent conditions II and III.
Figure 1.
Typical ATHP [M+2H]+2, [M+3H]+3 and [M+4H]+4 CCS profiles as a function of the ESI solvent conditions I) water:methanol:acetonitrile (33:33:33), II) water:methanol:acetic acid (49:49:2), and III) 10 mM ammonium acetate. Dashed lines are used to identify isomer bands.
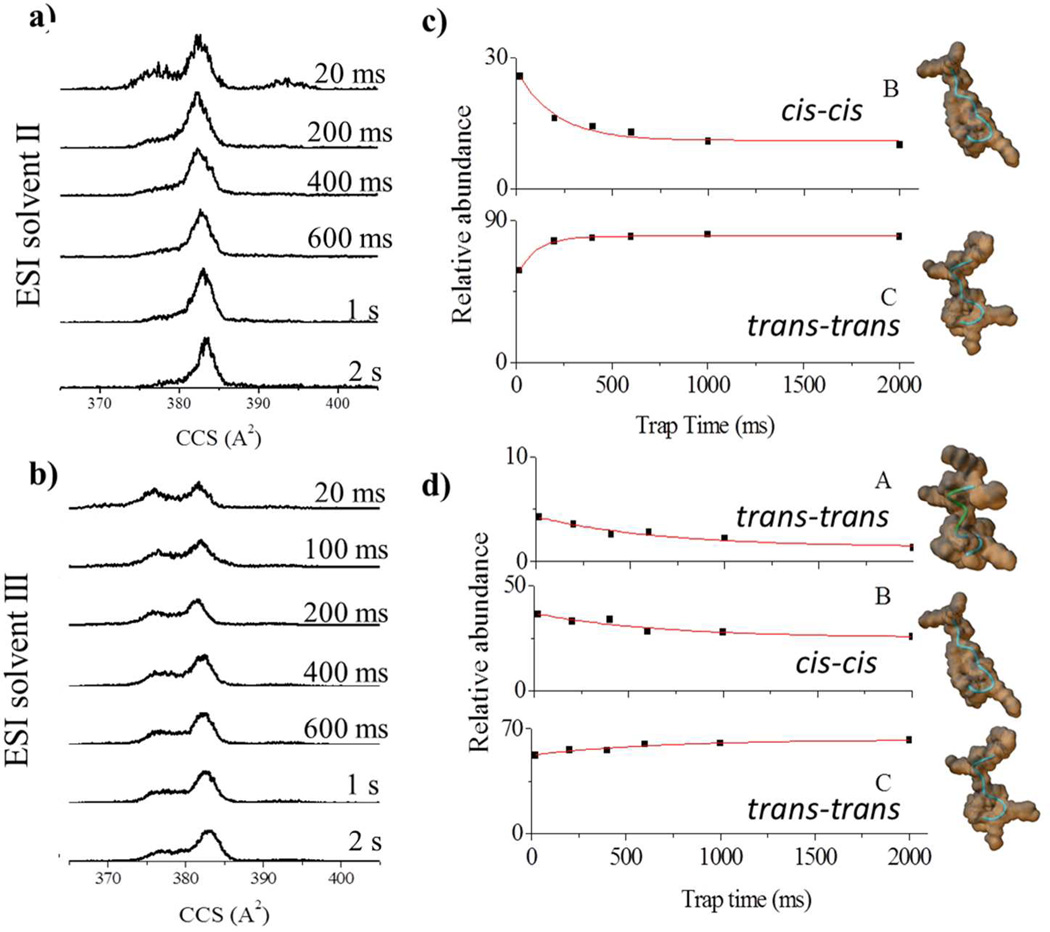
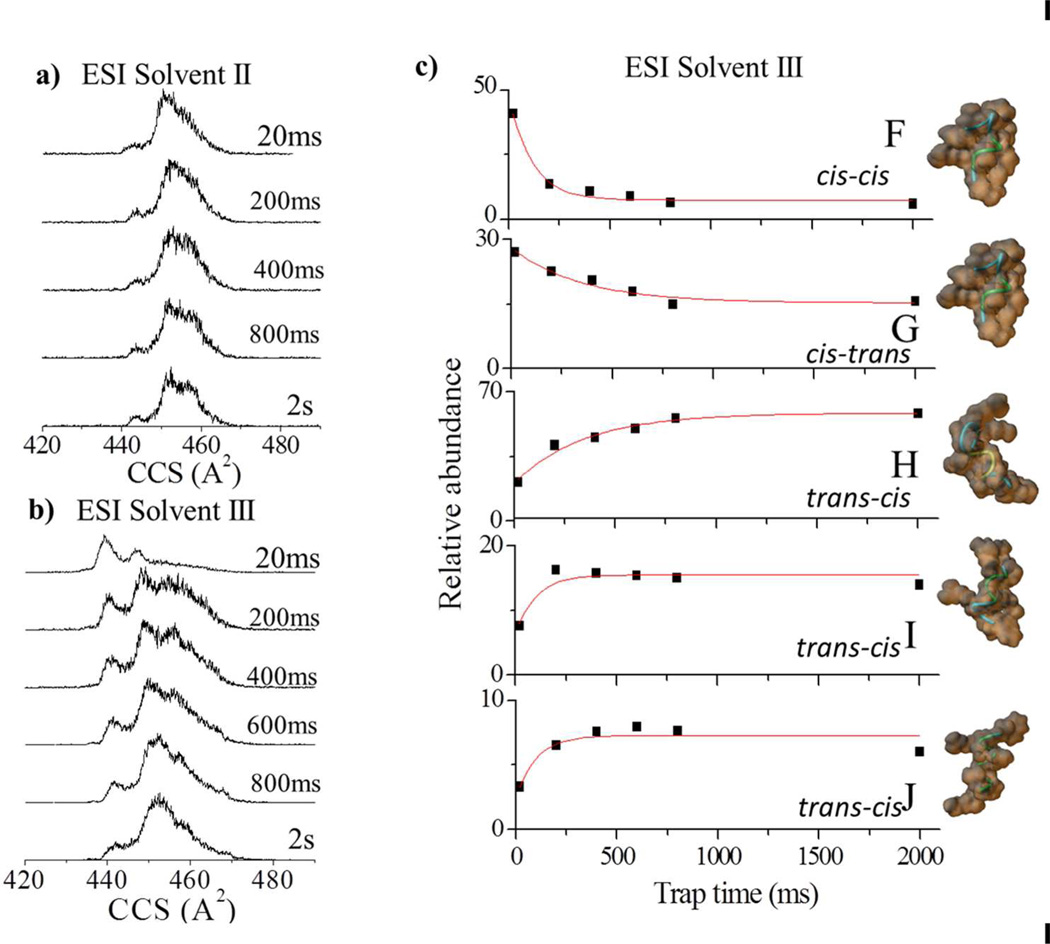
For the ATHP [M+2H]+2, the relative abundance of the IMS bands varied as a function of the trapping time and the ESI solvent conditions II-III (Figure 2). For ESI solvent condition II, the main pathway observed was B to C (150 ms conversion time). For ESI solvent condition III, the main pathway observed was A and B to C (650 ms growth time). Independent of ESI solvent condition, isomerization towards conformation C occurred, suggesting an overall higher stability for the “desolvated state” of ATHP [M+2H]+2 conformation C. For the ATHP [M+3H]+3, the relative abundance of the IMS bands varied as a function of the trapping time only for ESI solvent conditions III; that is, no variations were observed for ESI condition I and II (Figure 3). When analyzed as a function of the trapping time, two conformational inter-conversion pathways are observed: G to H (350 ms growth time) and F to J and I (100 ms growth time). We attribute the smaller variations for ATHP [M+3H]+3 to a lower flexibility of the conformational isomers at a higher charge state independent of the solution conditions. This behavior is consistent with the broad band observed for ATHP [M+4H]+4. That is, due to the small size of the molecule (decapeptide), intramolecular columbic repulsion (mainly between the Arg and Lys side chains) plays a larger role in the stabilization of the bulk molecular structure at higher charge states. In addition, heating of the conformational isomers was performed with the purpose of selectively annealing isomers into more stable forms by varying the ion effective temperature (i.e., RF amplitude 150–250 Vpp) during the trapping step in the TIMS cell. This approach is similar to collision activation experiments performed prior to IMS analysis27,28, and may lead to the observation of gas-phase quasi-equilibrium distribution states by changing the ion effective temperature22,29–31. For ESI solvent condition II and III, with the increase of the RF amplitude, the ATHP [M+2H]+2 and [M+3H]+3 charge state distributions shift to mainly C and H isomers, respectively. That is, the end step conformations obtained with the RF heating are the same that as those that predominate in the trapping experiments and more likely the more stable “desolvated” conformers.
Figure 2.
Typical ATHP [M+2H]+2 CCS profiles for ESI solvent conditions a) II and b) III as a function of the trap time (20 ms to 2 s). The relative abundances and candidate structures for the most abundant bands (A–C) as a function of the trap time is depicted in c) and d) for ESI solvent condition II and III, respectively. Experimental data have been fitted with exponential decays. Candidate structures are displayed on the far right with the orientations of the proline residues presented
Figure 3.
Typical ATHP [M+3H]+3 CCS profiles for ESI solvent conditions a) II and b) III as a function of the trap time (20 ms to 2 s). The relative abundances and candidate structures for the most abundant bands (F–J) as a function of the trap time is depicted in c) for ESI solvent condition III. Experimental data are fitted with exponential decays. Candidate structures are displayed on the far right with the orientations of the proline residues presented.
Complementary information was obtained by theoretically studying the conformational dynamics of ATHP in the TIMS conditions (i.e., collisions with nitrogen molecules at a bath temperature of ca. 300K). In particular, molecular dynamic simulations were used to investigate the inter-conversion dynamics as a function of the charge state and protonation site. From the simulations, candidate structures were proposed for the isomer bands observed for the ATHP [M+2H]+ and the [M+3H]+ distributions (Figures 2 and 3, respectively). The ATHP [M+2H]+2 conformational inter-conversion follows different mechanisms. That is, it can be a result of relaxation in the peptide backbone (inter-conversion of band A to D), re-orientation of side chains (inter-conversion of band E to D) and a combination of these two mechanisms (interconversion of band B to C). For ATHP [M+3H]+3, main differences between the candidate structures also arises from changes in the backbone, side-chain orientation and protonation site. Previous solution phase NMR experiments showed that ATHP binding does not significantly perturb the DNA conformation in the “AT-hook motif” and that all proline residues are in the trans conformation (similar to conformation C).11,32 The central RGR core deeply penetrates to the minor groove of AT base pairs and forms extensive electrostatic and hydrophobic contacts with the floor of the minor groove. The two prolines direct the motif away from the floor of the minor groove and place the positively charged amino acids near the negatively charged phosphate backbone making further contacts with DNA.11 However, a small population of peptides with cis-proline isomers was also observed in good agreement with the candidate structures proposed in Figures 2 and 3.32 The relative stability of the conformational isomers proposed in Figures 2 and 3 are also in good agreement with the NMR observations. That is, the all trans proline conformation is the more stable, and stability decreases with the increase of cisproline isomers. Similar backbone stabilization of the conformational space in proline containing peptides has been previously observed.22,23,28
CONCLUSIONS
In solution, ATHP conformations are expected to be selectively stabilized by the solvent conditions and by specific binding to the DNA portion. Previous observation of multiple structures16,32 and the results presented here provide a means to better describe the interaction dynamics of ATHP folding and ATHP desolvation. The results presented here also showed that TIMS-MS is a valuable tool to investigate solvent states and that isomerization kinetics can be followed at the level of side chain interaction and backbone relaxation. It is also shown that ATHP is “structured” and that ATHP conformations are defined by the protonation site, backbone and side orientations and can retain the memory of the initial solvent distribution during desolvation.
Supplementary Material
Table 1.
Reduced Mobility (Ko) and Collision Cross Section (CCS) values in nitrogen of ATHP [M+2H]+2, [M+3H]+3 and [M+4H]+4 observed as a function of the ESI solvent conditions.
| Isomer band | Ko (cm2 V−1s−1) |
CCS [A2] | ESI solution |
Decay ↓/ Growth ↑ Lifetime (ms) |
|
|---|---|---|---|---|---|
| ESI Solvent Condition II |
ESI Solvent Condition III |
||||
| [M+2H]+2 A | 1.0878 | 370 | III | ↓ 650±30 | |
| [M+2H]+2 B | 1.0699 | 376 | I, II, III | ↓ 150±40 | ↓ 650±30 |
| [M+2H]+2 C | 1.0543 | 382 | I, II, III | ↑ 150±50 | ↑ 650±30 |
| [M+2H]+2 D | 1.0311 | 390 | II | No variation | - |
| [M+2H]+2 E | 1.0241 | 393 | I, III | No variation | |
| [M+3H]+3 F | 1.3891 | 435 | I, II, III | - | ↓ 100±20 |
| [M+3H]+3 G | 1.3743 | 439 | I, II, III | - | ↓ 350±30 |
| [M+3H]+3 H | 1.3487 | 448 | I, II, III | - | ↑ 350±30 |
| [M+3H]+3 I | 1.3260 | 455 | I, II, III | - | ↑ 100±20 |
| [M+3H]+3 J | 1.3051 | 463 | III | - | ↑ 100±20 |
| [M+4H]+4 | 1.4718 | 547 | I, II, III | - | - |
ACKNOWLEDGMENT
This work was supported by National Institute of Health (Grant No. R00GM106414). The authors wish to acknowledge Dr. Desmond Kaplan from Bruker Daltonics Inc. for the development of TIMS acquisition software.
Funding Sources:
Funding has been provided by the National Institute of Health, Grant No. R00GM106414.
ABBREVIATIONS
- ATHP
AT-hook peptide
- CCS
collision cross section
- ESI
electrospray ionization
- IMS
ion mobility spectrometry
- TIMS-MS
trapped ion mobility mass spectrometry
Footnotes
ASSOCIATED CONTENT:
Supplementary information:
Supporting information includes the PDB files of the ATHP candidate structures proposed and a table containing the proline residue isomerization, protonation site and relative energy.
REFERENCES
- 1.Young AR, Narita M. Genes Dev. 2007;21:1005–1009. doi: 10.1101/gad.1554707. [DOI] [PubMed] [Google Scholar]
- 2.Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J, Chada K. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, Benson KF, Ashar HR, Chada K. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 4.Anand A, Chada K. Nat. Genet. 2000;24:377–380. doi: 10.1038/74207. [DOI] [PubMed] [Google Scholar]
- 5.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, St Pourcain B, Evans DM, Charoen P, Kaakinen M, Cousminer DL, Lehtimaki T, Kreiner-Moller E, Warrington NM, Bustamante M, Feenstra B, Berry DJ, Thiering E, Pfab T, Barton SJ, Shields BM, Kerkhof M, van Leeuwen EM, Fulford AJ, Kutalik Z, Zhao JH, den Hoed M, Mahajan A, Lindi V, Goh LK, Hottenga JJ, Wu Y, Raitakari OT, Harder MN, Meirhaeghe A, Ntalla I, Salem RM, Jameson KA, Zhou K, Monies DM, Lagou V, Kirin M, Heikkinen J, Adair LS, Alkuraya FS, Al-Odaib A, Amouyel P, Andersson EA, Bennett AJ, Blakemore AI, Buxton JL, Dallongeville J, Das S, de Geus EJ, Estivill X, Flexeder C, Froguel P, Geller F, Godfrey KM, Gottrand F, Groves CJ, Hansen T, Hirschhorn JN, Hofman A, Hollegaard MV, Hougaard DM, Hypponen E, Inskip HM, Isaacs A, Jorgensen T, Kanaka-Gantenbein C, Kemp JP, Kiess W, Kilpelainen TO, Klopp N, Knight BA, Kuzawa CW, McMahon G, Newnham JP, Niinikoski H, Oostra BA, Pedersen L, Postma DS, Ring SM, Rivadeneira F, Robertson NR, Sebert S, Simell O, Slowinski T, Tiesler CM, Tonjes A, Vaag A, Viikari JS, Vink JM, Vissing NH, Wareham NJ, Willemsen G, Witte DR, Zhang H, Zhao J, Wilson JF, Stumvoll M, Prentice AM, Meyer BF, Pearson ER, Boreham CA, Cooper C, Gillman MW, Dedoussis GV, Moreno LA, Pedersen O, Saarinen M, Mohlke KL, Boomsma DI, Saw SM, Lakka TA, Korner A, Loos RJ, Ong KK, Vollenweider P, van Duijn CM, Koppelman GH, Hattersley AT, Holloway JW, Hocher B, Heinrich J, Power C, Melbye M, Guxens M, Pennell CE, Bonnelykke K, Bisgaard H, Eriksson JG, Widen E, Hakonarson H, Uitterlinden AG, Pouta A, Lawlor DA, Smith GD, Frayling TM, McCarthy MI, Grant SF, Jaddoe VW, Jarvelin MR, Timpson NJ, Prokopenko I, Freathy RM. Nat. Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, Mader H, Kuchenbauer F, Humphries RK, Eaves CJ. Nat. Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 8.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga JJ, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O'Brien C, Papmeyer M, Putz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TG, van Tol MJ, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Goring HH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW, Jr, Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Muhleisen TW, Nauck M, Nothen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Renteria ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdes Hernandez MC, Van den Heuvel M, van der Wee NJ, Van Haren NE, Veltman JA, Volzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR, Jr, Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW, Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S, Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJ, Espeseth T, Fernandez G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer-Lindenberg A, Morris DW, Muller-Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Samann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM. Nat. Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui T, Leng F. Biochemistry. 2007;46:13059–13066. doi: 10.1021/bi701269s. [DOI] [PubMed] [Google Scholar]
- 10.Lehn DA, Elton TS, Johnson KR, Reeves R. Biochem. Int. 1988;16:963–971. [PubMed] [Google Scholar]
- 11.Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 12.Reeves R. Biochem. Cell Biol. 2003;81:185–195. doi: 10.1139/o03-044. [DOI] [PubMed] [Google Scholar]
- 13.Reeves R. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 14.Fenn J, Mann M, Meng C, Wong S, Whitehouse C. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 15.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 16.Tao L, McLean JR, McLean JA, Russell DH. J. Am. Soc. Mass Spectrom. 2007;18:1232–1238. doi: 10.1016/j.jasms.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Valentine SJ, Counterman AE, Clemmer DE. J. Am. Soc. Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 18.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Nat. Protocols. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Lima FA, Blase RC, Russell DH. Int. J. Mass Spectrom. 2010;298:111–118. doi: 10.1016/j.ijms.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beveridge R, Chappuis Q, Macphee C, Barran P. Analyst. 2013;138:32–42. doi: 10.1039/c2an35665a. [DOI] [PubMed] [Google Scholar]
- 21.Wyttenbach T, von Helden G, Bowers MT. J. Am. Chem. Soc. 1996;118:8355–8364. [Google Scholar]
- 22.Fernandez-Lima FA, Wei H, Gao YQ, Russell DH. J. Phys. Chem. A. 2009;113:8221–8234. doi: 10.1021/jp811150q. [DOI] [PubMed] [Google Scholar]
- 23.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. J. Am. Chem. Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Lima FA, Kaplan DA, Suetering J, Park MA. Int. J Ion Mobil. Spectrometry. 2011;14:93–98. doi: 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Lima FA, Kaplan DA, Park MA. Rev. Sci. Instr. 2011;82:126106. doi: 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.United States. 1999 [Google Scholar]
- 27.Pierson NA, Valentine SJ, Clemmer DE. J Phys. Chem. B. 2010;114:7777–7783. doi: 10.1021/jp102478k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson NA, Chen L, Russell DH, Clemmer DE. J Am. Chem. Soc. 2013;135:3186–3192. doi: 10.1021/ja3114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wysocki VH, Kenttämaa HI, Cooks RG. Int. J Mass Spectrom. and Ion Processes. 1987;75:181–208. [Google Scholar]
- 30.Donald WA, Khairallah GN, O'Hair RA. J Am. Soc. Mass. Spectrom. 2013;24:811–815. doi: 10.1007/s13361-013-0625-x. [DOI] [PubMed] [Google Scholar]
- 31.Robinson EW, Shvartsburg AA, Tang K, Smith RD. Anal. Chem. 2008;80:7508–7515. doi: 10.1021/ac800655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geierstanger BH, Volkman BF, Kremer W, Wemmer DE. Biochemistry. 1994;33:5347–5355. doi: 10.1021/bi00183a043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.