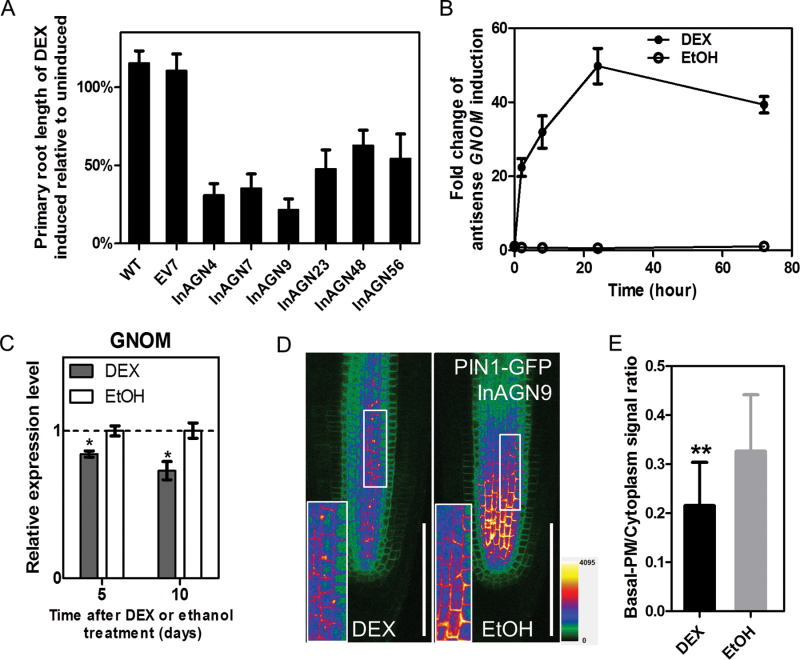
Fig. 1.
Transient suppression of GNOM expression in Arabidopsis and phenotypes of seedlings. (A) Primary root length of DEX-induced seedlings relative to uninduced seedlings in different transgenic lines grown on DEX- or EtOH-supplemented MS medium for 7 d after germination. The EV7 line contains empty vector with no antisense construct. The mean ±SE for 24 seedlings in triplicate experiments is plotted. (B) Fold change of the induced GNOM antisense transcripts after InAGN9 seedlings were exposed to DEX or EtOH for various times. EtOH indicates the uninduced control. Each data point represents the mean ±SE from three biological repeats and two technical repeats. (C) Expression of endogenous GNOM mRNA in induced or uninduced InAGN9 seedlings. Each data point represents the mean ±SE from two biological repeats and two technical repeats. *P<0.05 (Student’s t-test). (D) Localization of GFP-fused PIN1 protein in root tip treated with DEX or EtOH. Insets: enlargements of boxed areas, showing details of PIN1 subcellular localization. (E) Fluorescent intensity of PIN1–GFP in the basal plasma membrane relative to that in the cytoplasm. DEX, n=30 cells from three roots; EtOH, n=52 cells from thee roots; **P<0.01 (Student’s t-test). Scale bar=100 μm in (D).