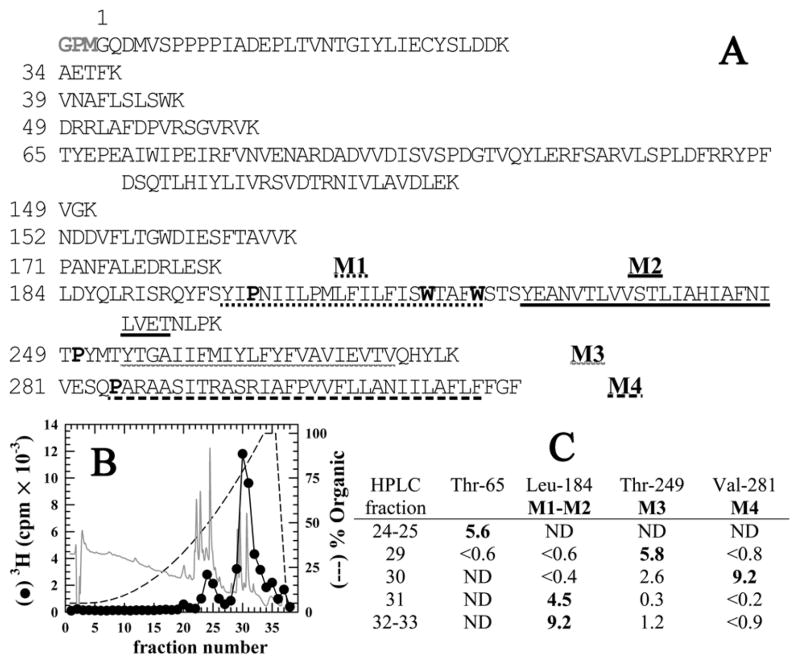
Figure 2. Reversed-phase HPLC fractionation of EndoLys-C digests of GLIC: [3H]AziPm primarily photoincorporate into the hydrophobic, transmembrane domain of GLIC.
A) Shown are the sequences of the GLIC fragments produced by enzymatic cleavage with EndoLys-C (specific for Lys). The residue numbering is that used in the crystal structure (PDB:3P50 (10)) and does not include the 3 additional N-terminal residues (GPM) identified by sequence analysis of intact, expressed GLIC. The transmembrane helices M1–M4 are underlined. The Pro residues used to chemically isolate M1, M3, & M4 during sequencing are bolded as are the Trp residues subjected to chemical cleavage to sequence M2. B) Reversed-phase HPLC fractionation of an EndoLys-C digest of [3H]AziPm labeled GLIC. 82% of the recovered 3H eluted in hydrophobic fractions (>75 % organic, fractions 28–38). In gray is the absorbance profile at 215 nm. C) Selected fractions containing 3H were sequenced, with the peptides detected quantitated in picomoles. In fractions 29–33, the only GLIC fragments detected were those containing the transmembrane helices. ND, not detected.