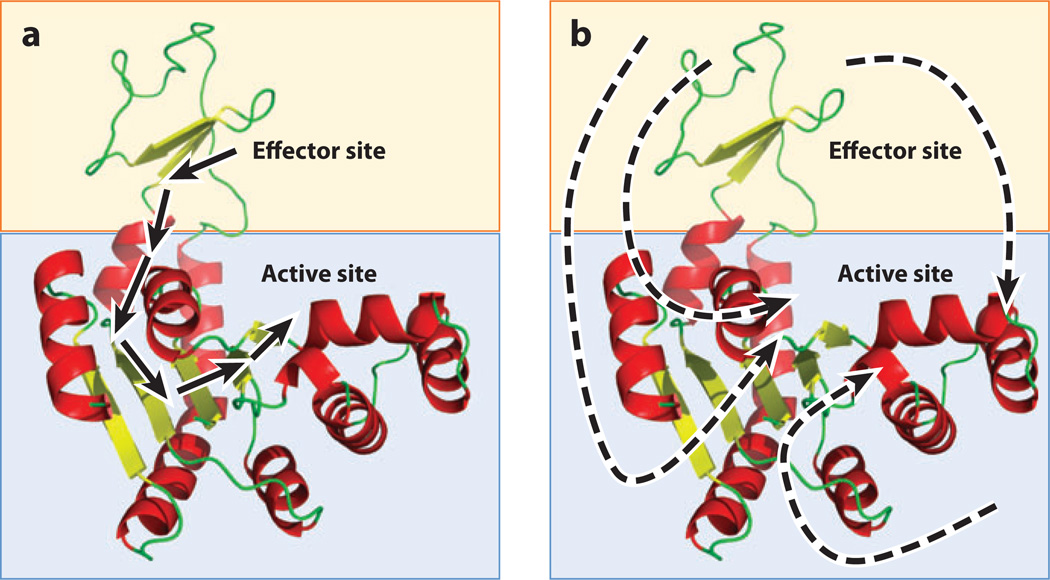
Figure 2.
The structure-based and non-structure-based views of allostery. (a) A hypothetical two-domain protein (blue and orange boxes) contains effector and active sites. The structure-based view of allostery posits that one or a small number of unique pathways of structural deformations (arrows) propagate energy between the sites, thus enabling the allosteric mechanism. Such a view tacitly assumes that the free energy of the allosteric mechanism is proportional to the enthalpy of the noncovalent bonds made and broken along this pathway. (b) A hypothetical two-domain protein (blue and orange boxes) contains effector and active sites. The non-structure-based view of allostery posits that regional changes in dynamics, protein, and/or solvent conformational entropy, or population shifts, are the dominant contributors to the free energy of the allosteric mechanism. Unique energetic pathways are difficult or impossible to place on a single molecular structure of the protein because the free energy is potentially a combination of enthalpic and entropic contributions from many sources.