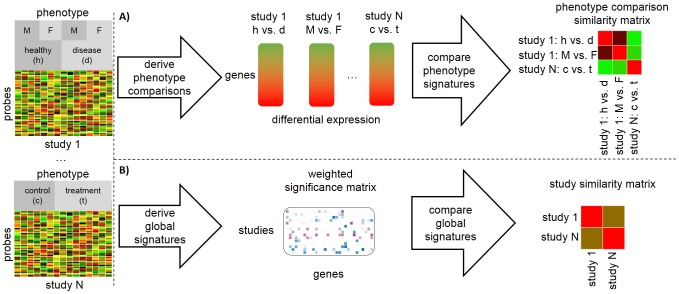
Figure 1. Summary of the steps performed (A) in the differential expression meta-analysis paradigm and (B) in our proposed framework.
Two studies from a collection of  independent studies are shown. One of the studies includes experimental annotations about gender (Male or Female) and disease status (Healthy or Disease), while the other study includes experimental annotations indicating whether each sample comes from a Control or a Treatment. In the differential expression approach each study is broken down into a set of comparisons between phenotypes, with the first study yielding two comparisons and the second study yielding a single comparison; genes are ordered according to a differential expression measure, e.g., log-ratios, which are used as a basis for computing the similarities between phenotype comparisons. In the proposed approach, one first decides if the analysis is based on absolute expression or differential expression. Then, in the case that one is interested in a differential expression analysis, the expression in every non-control sample (e.g. Treatment or Disease) is computed relative to the corresponding control sample. Afterwards, the rank product method is used to compute global, study-wide features corresponding to genes that possess a consistently high expression or differential expression in a given study. These global features are statistically modelled and used as a basis for deriving similarities between studies. The connections are therefore established not between subsets of studies, but rather directly between studies.
independent studies are shown. One of the studies includes experimental annotations about gender (Male or Female) and disease status (Healthy or Disease), while the other study includes experimental annotations indicating whether each sample comes from a Control or a Treatment. In the differential expression approach each study is broken down into a set of comparisons between phenotypes, with the first study yielding two comparisons and the second study yielding a single comparison; genes are ordered according to a differential expression measure, e.g., log-ratios, which are used as a basis for computing the similarities between phenotype comparisons. In the proposed approach, one first decides if the analysis is based on absolute expression or differential expression. Then, in the case that one is interested in a differential expression analysis, the expression in every non-control sample (e.g. Treatment or Disease) is computed relative to the corresponding control sample. Afterwards, the rank product method is used to compute global, study-wide features corresponding to genes that possess a consistently high expression or differential expression in a given study. These global features are statistically modelled and used as a basis for deriving similarities between studies. The connections are therefore established not between subsets of studies, but rather directly between studies.