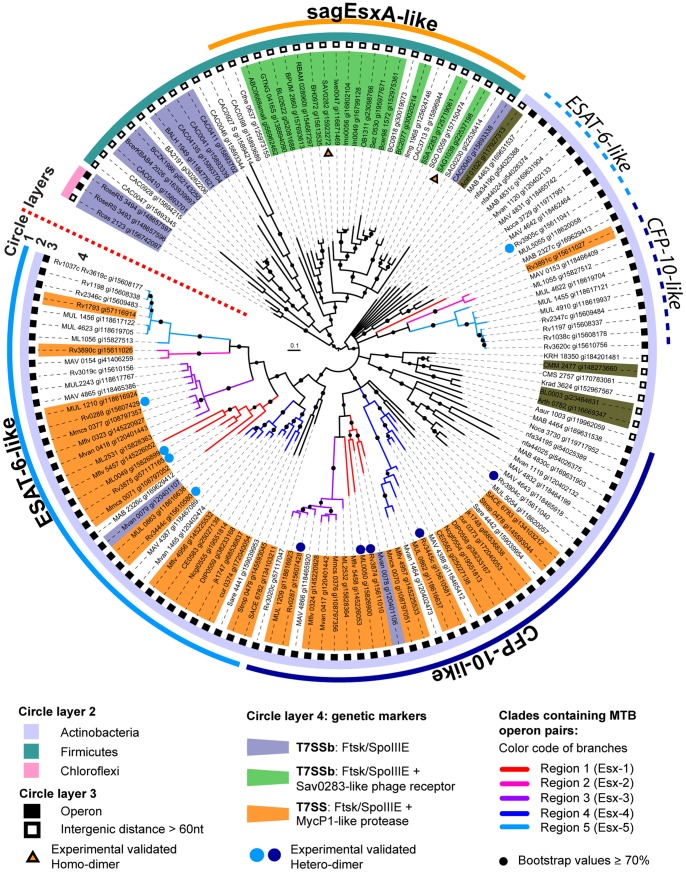
Figure 2. Estimated phylogenetic tree of the WXG-100 protein family consisting of three WXG100 subfamilies.
The tree of WXG100 proteins was constructed in midpoint rooted presentation with three main clades: CFP-10-like (blue circular arc), ESAT-6-like (cyan circular arc) proteins and sagEsxA-like proteins (orange circular arc). The WXG100 gene pairs of M. tuberculosis occurring within the RD1-like gene clusters denoted as the regions (Esx) 1 to 5 are coloured accordingly along with the Rv-annotations (see subtitles). The annotations of the genes in close proximity to each of the WXG100 genes were manually analyzed and this information was also included to the tree. Two WXG100 genes with an intergenic distance of less than 80 nucleotides (according to the definition Roback et al. [47]) are considered to be encoded within a bi-cistronic operon (filled black squares on the circle layer 3), whilst mono-cistronic WXG100 genes are indicated by an unfilled squares. Those WXG-proteins whose oligomeric properties have been experimentally determined are marked with a triangle for homodimers and with pairs of blue dots for heterodimers. The second inner arcs show the phyla of the bacteria.