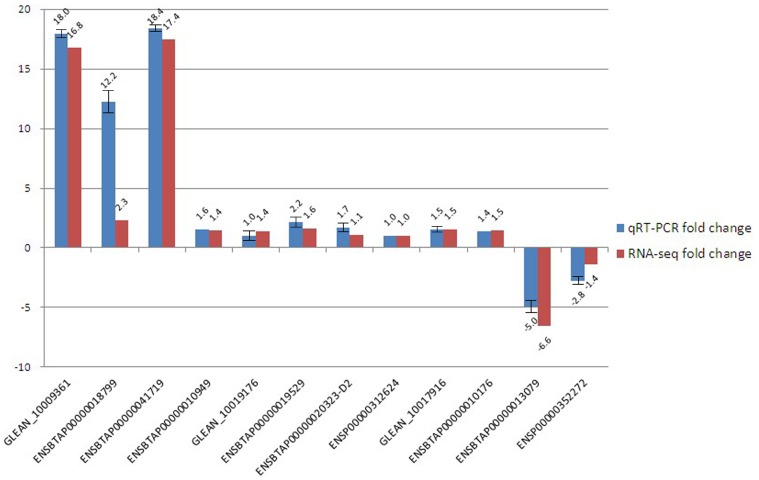
Figure 3. qRT-PCR validation of the expressed genes using the Illumina sequencing technology.
For the 12 randomly selected differentially expressed genes, fold changes of DP/SH determined from the relative Ct values of using the 2−△△CT method in qRT-PCR were compared to those detected by RPKM of DP/SH in RNA-seq. All Ct values were normalized to GAPDH and replicates (n = 3) of each sample were run.