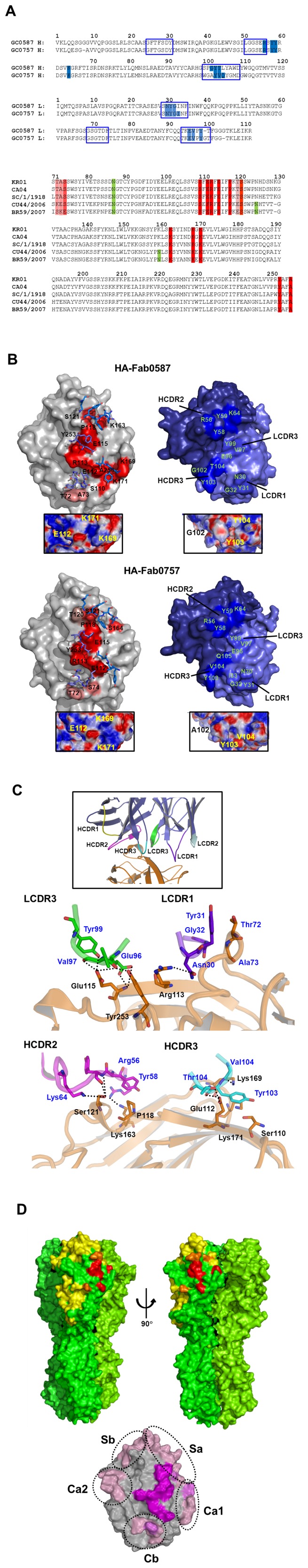
Figure 3. Sequence alignments and antigenic sites.
(A) Sequence alignments of variable region of GC0587 and GC0757 (upper panel) and H1 HAs (lower panel). Residues in CDRs are in blue open boxes and residues that interact with HA are highlighted in blue filled boxes. Epitopes in H1 HAs are highlighted in pink, and more conserved residues are highlighted in red. Potential glycosylation sites are highlighted in green. (B) Surface representations of KR01 HAs (gray) and Fab fragments (dark blue and light blue for H-chain and L-chain, respectively). Antigenic sites are colored in red for highly conserved residues and pink for moderately conserved residues. Amino acid residues involved in the interactions between KR01 HA and Fab0587 are colored in blue and slate. Insets are surface charge representations with contours from −10 (red) to+10 (blue) kT through 0 (white). (C) Detailed interactions of HA with Fab0587. HA and Fab are colored in orange and blue, respectively. Residues that contribute to the interactions are represented as stick models. LCDR1 and LCDR3 are colored in purple and green, respectively (upper panel), and HCDR2 and HCDR3 are colored in magenta and cyan, respectively (lower panel). (D) Residues found at other structurally characterized antibody complexes are colored in yellow, those at both Fab0587 and other antibody complexes are in orange, and those against GC0587 are in red. Classical antigenic sites are colored in light pink (Ca, Cb, Sa, and Sb) and those at both Fab0587 antigenic sites are in pink.