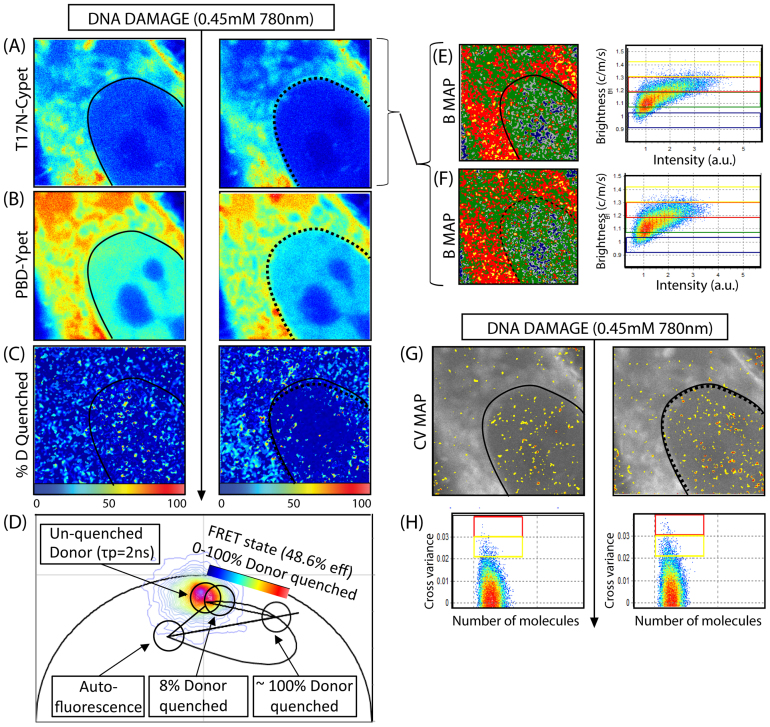
Figure 3. Inactive mutant of Rac1 (T17N) FRET biosensor activity and oligomerisation during the DNA damage response.
(A)–(B) Intensity image of a NIH3T3 nucleus transiently transfected with T17N-CyPet (donor) and PBD-YPet (acceptor) respectively, before (left) and immediately after (right) induction of DNA damage. (C) FLIM images of the biosensor before (left) and after (right) DNA damage depicted in (A)–(B) and pseudo-colored according to the palette defined in (D), which spans the 0-100% blue to red range of donor quenching that is projected for the Rac1 biosensor FRET efficiency of 48.6%. As can be seen from this representation, after induction of DNA damage the inactive mutant T17N completely inhibited nuclear activation of Rac1 but resulted in activation of 8% of cytoplasmic Rac1. (D) The phasor distribution of lifetimes measured for the T17N biosensor experiment depicted in (A)–(B), with the theoretical FRET trajectory superimposed with respect to the unquenched donor (tau phase 2 ns) and background (auto-fluorescence). (E)–(F) Molecular brightness of T17N-CyPet before (E) and after (F) DNA damage pseudo-coloured according to the coloured cursors superimposed over the brightness versus intensity distributions (right): monomers coloured green, dimers coloured red, higher order oligomers coloured yellow. Dimers are excluded from the nucleus and this localisation does not change upon induction of DNA damage. (G) Cross variance analysis of biosensor FRET experiment depicted in (A)–(B) pseudo-colored according to the palette defined in (H). (H) Cross variance analysis of T17N-CyPet intensity fluctuation with PBD-YPet intensity fluctuation recorded in each pixel of the image. In (A–C), (F) and (G), nuclear shape before and after DNA damage are indicated by solid and dashed black lines, respectively.