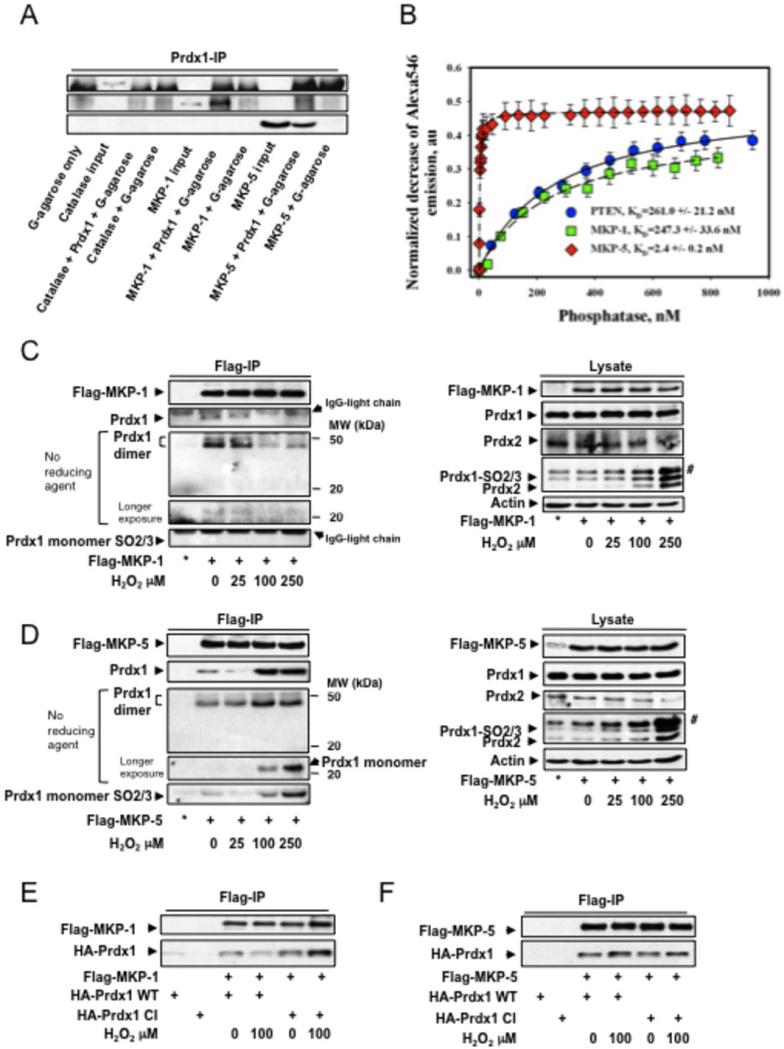
Figure 3. Non-covalent binding of Prdx1 to MKP-1 and MKP-5.
A. Pull down of recombinant Catalase, MKP-1 and 5 by Prdx1 conjugated protein G agarose. The far left lane represents protein agarose G only to serve as a negative control for non-specific binding. B. Purified Prdx1 protein (2.0 nM) labeled with Alexa Fluor® 546 was titrated with indicated amount of the purified phosphatase proteins labeled with QSY® 35. Alexa546 fluorescence decrease (normalized to initial Alexa546 emission) was recorded and processed using a “One Site Saturation model” (Pharmacology application, SigmaPlot 10.0, SyStat, MA) with best hyperbolic fit (R2≥ 0.99) according to equation: Y= Bmax*X / (KD + X) where: X – is a concentration of added PTEN (MKP-1 or MKP-5) protein; Y – is a normalized decrease of Alexa®546 fluorescence corresponding to specific binding of phosphatases; Bmax – is a saturated number of binding sites with apparent equilibrium dissociation constant KD. Data represent mean±SD for 3 independent experiments. The lines represent hyperbolic fit of experimental data for the titration of 2.0 nM of Alexa546-labeled Prx1 with QSY35-labeled: PTEN – solid line (R2=0.99); MKP-1 - dashed line (R2=0.99); and MKP-5 - doted and dashed line (R2=0.99). C and D. 293T HEK cells were transfected with 2.0μg of Flag-MKP-1 and Flag-MKP-5, and treated with increasing amounts of H2O2 for 30 min in serum free medium. To assay for binding of endogenous Prdx1, 1000 μg of protein lysate was immunoprecipitated using anti-flag affinity matrix and incubated 3 hrs at 40C in an hypoxic chamber. The affinity matrix was harvested by centrifugation at 3000xg for 2 min, and washed with 0.5 ml of lysis buffer 3 times. The resin was re-suspended in 20 μl of reducing or non-reducing SDS-PAGE sample buffer, boiled 10 mins, and analyzed by Western blotting for MKP expression, Prdx1 and Prdx1Cys52SO3 binding. * = IP from untransfected cells. Co-IPs were also analyzed in the absence of β-mercaptoethanol. No Prdx1 dimer staining positive for SO2/3 were detected. #: detection of a higher molecular weight 2-Cys Prdx family member. Co-IP unbound fraction can be found in the supplemental material Fig. S3B. E and F. 293T HEK cells were cotransfected with Flag-MKP-1 or Flag-MKP-5 either with HA-Prdx1WT or HA-Prdx1CI, and treated with increasing amounts of H202 for 30 min in serum free medium. For co-IP, 1000μg of lysate was added to anti-flag affinity matrix and incubated for 1h at 40C. Following incubation, the affinity matrix was harvested as stated above, re-suspended in 20μl of reducing sample buffer, and boiled for 10 min. Binding of HA-Prdx1WT and HA-Prdx1CI was determined by Western blotting. Analysis of cell lysate for protein expression can be found in the supplemental material Fig. S3C.