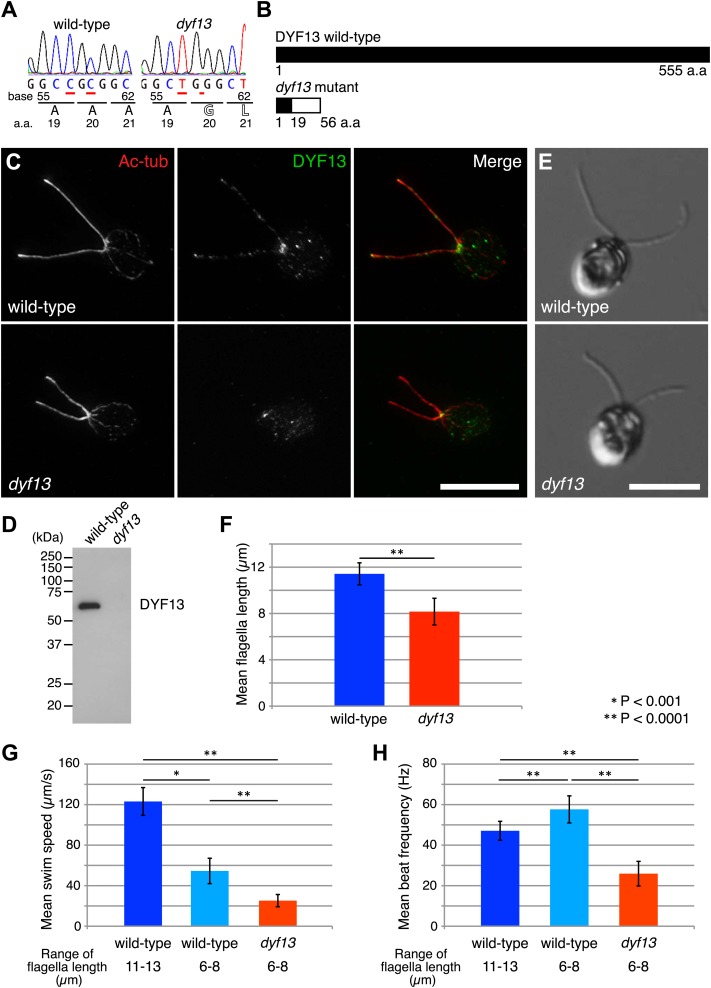
Figure 3. C. reinhardtii dyf13 mutant has short flagella and motility defects.
(A) Sequences of the DYF13 gene (RefSeq accession: XM_001698717) in wild-type (CC125) and dyf13 mutant cells. Red underlines show places of mutations. A one-base substitution (c.57C >T) does not change amino acid. A one-base deletion (c.59delC) in the dyf13 gene induces frame-shift after 20th amino acid (a.a.) reside (p.Ala20Glufs). (B) Schematic representation of C. reinhardtii DYF13 (XP_001698769) and its mutant. The black bar indicates correct amino acid sequence and the white bar indicates frame-shift sequence. (C) Immunofluorescence images of wild-type and dyf13 mutant cells. The cells were stained with antibodies to acetylated tubulin (red) and DYF13 (green). Bar: 10 μm. (D) Isolated flagella fractions from wild-type cc125 and dyf13 mutant cells were analyzed in Western blots probed with the DYF13 antibody. The DYF13 antibody specifically recognized a ∼56 kDa band. (E) DIC images of wild-type and dyf13 mutant cells. Bar: 10 μm. (F–H) Flagella length (F), swim speed (G), and beat frequency of flagella (H) of wild-type (blue, n = 12) and dyf13 mutant cells (red, n = 12, p<1 × 10−6, 10−8, 10−12, respectively). Swim speeds and beat frequencies of flagella were measured also in wild-type cells with short flagella, which were adjusted by pH shock and regeneration (light blue, n = 10). Error bars represent standard deviations.