Abstract
Hepatocellular carcinoma (HCC) represents one of the leading causes of cancer death and has proved to be highly refractory to treatment. Extensive analysis of the disease has demonstrated that it arises predominantly in response to high-risk etiological challenges, most notably hepatitis virus. However, with evolving vaccination and the obesity epidemic, progressively more cases are associated with underlying metabolic dysfunction. Pathologically diverse forms of HCC are observed, and recent sequencing analysis has defined common events that target well-known cancer pathways including β-catenin/Axin, TP53, and RB/CDKN2A, as well as frequent aberrations in chromatin remodeling factors. However, there are a myriad of low frequency genetic events that make each HCC case unique. Gene expression profiling approaches have successfully been deployed for prognostic assessment of hepatocellular carcinoma and to detect the earliest stages of disease. Despite more extensive research, systemic treatment for HCC is exceedingly limited, with only a handful of drugs providing benefit. Ongoing clinical trials are attempting to exploit specific biological dependencies of HCC to improve the dismal prognosis. Overall, the future of HCC treatment will rely on an understanding of the interplay between etiological factors, molecular features of disease, and rational therapeutic intervention.
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is the third leading cause of cancer-related death worldwide.1 It is the fifth most common cause of cancer in men (7.9% of all cancers) and the seventh most common cancer in women (6.5% of all cancers).2 There is considerable geographic variation in HCC incidence, with the majority of cases occurring in developing countries. More than 75% of HCC occurs in Southeast Asia and sub-Saharan Africa, with incidence rates exceeding 20 per 100,000 individuals. Southern European countries have intermediate incidence rates, whereas the lowest incidence rates (<5 per 100,000 individuals) are found in North America, South America, and Northern Europe.2
Etiological Factors
Hepatitis B virus (HBV) is the primary etiological factor for HCC worldwide, particularly in high-incidence areas, such as Asia and Africa, where more than 70% of HCC patients have underlying HBV infection.3 Annual incidence among HBV carriers from Asia exceeds 0.2% starting at age 40; therefore, HCC surveillance is recommended in Asian males older than age 40 and Asian females older than age 50, even in the absence of cirrhosis.4 Patients from Africa are particularly at high risk, potentially related to a synergistic effect from aflatoxin exposure, and experts recommend HCC surveillance at an earlier age.5 HBV-infected patients who are exposed to aflatoxin are 60 times more likely to develop HCC compared to those with neither exposure, which is likely related to HBV modulating the extent that aflatoxins can bind to DNA and result in mutations in the TP53 tumor-suppressor gene.6
In Europe and the United States, hepatitis C virus (HCV)-associated cirrhosis is the most common etiological factor, accounting for approximately 60% of HCC cases.7 The risk of HCC is increased 17-fold in HCV-infected patients compared to HCV-negative patients.8 However, HCC risk among patients with chronic HCV infection is primarily limited to those with cirrhosis, with an annual incidence rate of 2% to 8%.9,10 Alcoholic cirrhosis is another well-recognized risk factor for HCC, and alcoholic liver disease has been reported to be a contributing factor in nearly one-third of all HCC cases worldwide.11,12 However, HCC incidence rates among patients with alcoholic cirrhosis may be overestimated, given that several studies linking alcoholic cirrhosis and HCC predated routine testing for HCV infection.
Nonalcoholic steatohepatitis (NASH) cirrhosis is anticipated to be the major etiological factor for HCC in the future, as the number of NASH cases continues to increase in parallel with the obesity and diabetes epidemics.13 Several studies support an association between NASH and an increased risk of HCC, although this risk appears to be limited primarily to those with cirrhosis. Patients with NASH, but no cirrhosis, typically have cumulative HCC mortality rates of <1%, whereas those with NASH-related cirrhosis have cumulative HCC incidence rates of 2.4% to 12.8%.14
Most recent reports suggest that the incidence of HCC in high- and intermediate-incidence areas may be stabilizing or falling.2 In China and Taiwan, this is related to more widespread HBV vaccination programs and higher rates of HBV treatment.15 In Japan and Southern Europe, the decrease in HCC incidence may relate to an aging cohort of patients with HCV infection.16,17 In contrast, the incidence of HCC in low-incidence areas, such as the United States, is rapidly increasing. In the 10-year period from 1995 to 2004, HCC had the largest increase in incidence among all solid tumors in the United States.18 The rising incidence of HCC in the United States is related to several factors, but the two most important are growing populations of patients with advanced HCV infection and NASH.2 In parallel with its rising incidence rate, HCC also has one of the fastest growing death rates among solid tumors. Whereas the prognosis for most solid cancers improved over the 10-year period from 1994 to 2003, the mortality rate for HCC nearly doubled.7,18
Surveillance and Early Detection
Because many patients with HCC are asymptomatic when tumors are at an early stage, routine surveillance in patients with cirrhosis is important (Table 1). Although predictive risk models have been created to better define a high-risk subgroup among cirrhotic patients, none have been externally validated or ready for routine use in clinical practice.19,20 Ultrasound is the most widely used radiological test for HCC surveillance.21 The sensitivity of ultrasound for early stage HCC is only 63% (95% CI 49% to 76%), meaning more than one-third of all tumors are missed or diagnosed at advanced stages.22 Although α fetoprotein (AFP) was included as an adjunct surveillance test to ultrasound in prior guidelines, there has been increasing debate regarding its usefulness in surveillance.23–27 Recent data suggest that AFP may improve the effectiveness of surveillance for detecting early HCC in clinical practice, increasing sensitivity from <50% for ultrasound or AFP alone to nearly 65% when the two tests are combined.26 Other tumor biomarkers that have been evaluated include des-γ-carboxy prothrombin and the lens culinaris-agglutinin reactive fraction of AFP (APF-L3%)28; however, a large multicenter study demonstrated that AFP may be more sensitive for early stage HCC than either of these two biomarkers.29 Glypican 3,30,31 GP73,32 osteopontin,33 squamous cell carcinoma antigen,34 human hepatocyte growth factor,35 and insulin growth factor-1,36 are examples of other biomarkers that are being evaluated, but data are preliminary and require validation in large cohorts. The field of proteomics may also provide tools to determine other novel serum biomarkers in HCC in the future.37
Table 1.
Patient Populations in Whom Surveillance Is Recommended
| Surveillance Recommended |
| Asian male hepatitis B carrier older than age 40 |
| Asian female hepatitis B carrier older than age 50 |
| African Blacks with hepatitis B |
| Hepatitis B carrier with family history of HCC |
| Cirrhosis from any etiology |
| Surveillance Benefits Uncertain |
| Asian hepatitis B carriers younger than age 40 (males) or younger than age 50 (females) |
| Hepatitis B carriers who contracted infection via horizontal transmission |
| Hepatitis C carriers without cirrhosis |
| Nonalcoholic fatty liver disease patients without cirrhosis |
Pathological Features of HCC and Clinical Prognostic Tools
Effective radiological methods can yield a diagnosis of HCC; however, pathological evaluation remains a key aspect of clinical management. Microscopically, HCC tends to resemble normal hepatocytes, depending on the degree of differentiation, and architecture within the tumor can recapitulate normal liver architecture in the early stages, mimicking normal liver cell plates. Most commonly, HCC produces acinar, pseudoglandular, trabecular, and solid growth patterns, often with multiple growth patterns within a single tumor (Figure 1). As the tumor progresses, the preservation of architecture is lost, with crowding and high nuclear to cytoplasmic ratio. When HCC arises in a background of cirrhosis, distinguishing characteristics of the underlying disease are not present sometimes, which is due to altered landmarks and vascular relationships; however, histological changes in the surrounding cirrhotic liver can often provide a clue to the etiology of the cirrhosis and HCC. NASH and alcoholic liver disease have several histological similarities, including steatosis, ballooning degeneration of hepatocytes, Mallory-Denk bodies, pericellular fibrosis, and mixed lobular inflammation. Typical histological findings of HCV infection include portal lymphoplasmacytic inflammatory infiltrate, occasionally with prominent lymphoid aggregates, and variable lobular inflammation. Histological findings of HBV can overlap with HCV, but ground glass hepatocytes are a distinctive histological feature that can help distinguish cases of HBV infection. Several recent studies have described a steatohepatitic variant of HCC that can be seen in patients with HCC related to steatohepatitis (NASH and/or alcoholic liver disease).38–41 This HCC variant is described as having steatohepatitis-like features within the tumor, including steatosis, ballooning degeneration of the malignant hepatocytes, intratumoral pericellular fibrosis, Mallory-Denk bodies, and inflammation (Figure 2). In a study by Salomao et al,40 36% of patients who developed HCC in the setting of steatohepatitis were diagnosed as having a steatohepatitic variant of HCC as compared to 1.3% of HCC patients without steatohepatitis. Immunohistochemically, there was diffuse loss of cytoplasmic CK8/18 and an increased number of activated hepatic stellate cells with the steatohepatitic HCC, identical to the pattern seen in the surrounding nonneoplastic liver.40
Figure 1.
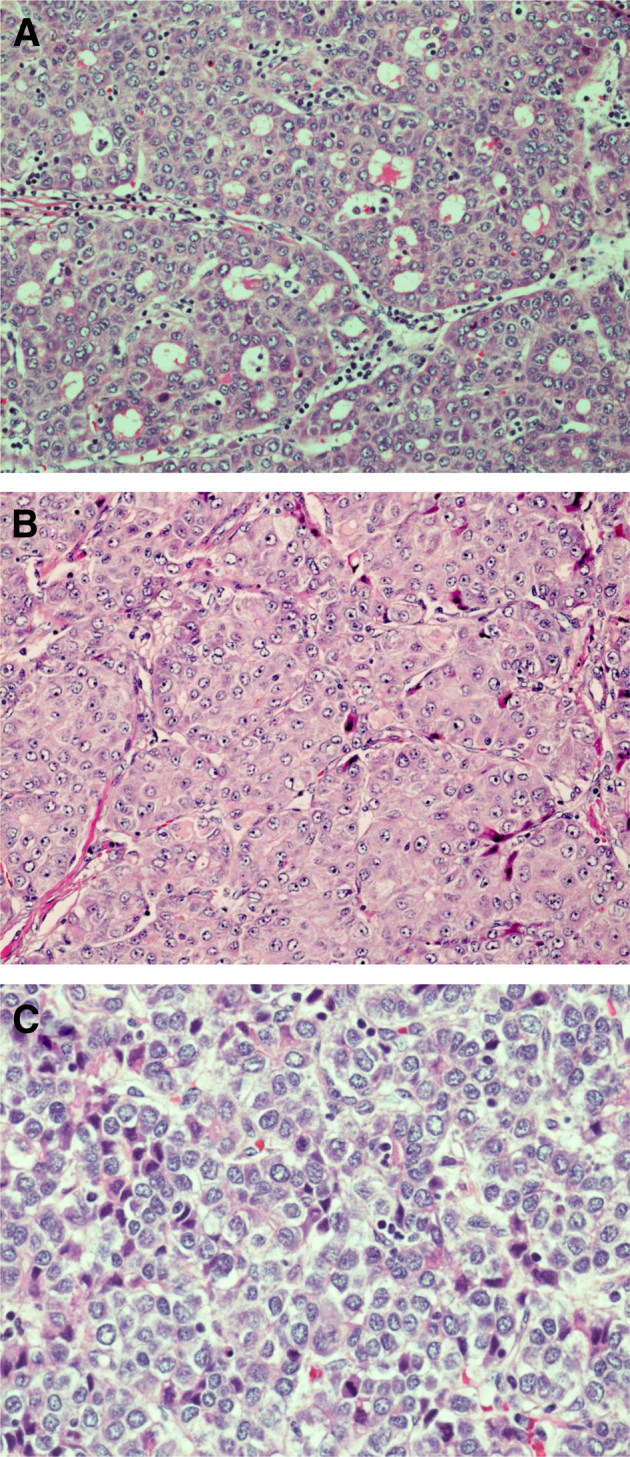
A: H&E staining for hepatocellular carcinoma, pseudoglandular growth pattern. B: Hepatocellular carcinoma, trabecular growth pattern. C: Hepatocellular carcinoma, solid growth pattern. Original magnification: ×20 (A); ×40 (B and C).
Figure 2.
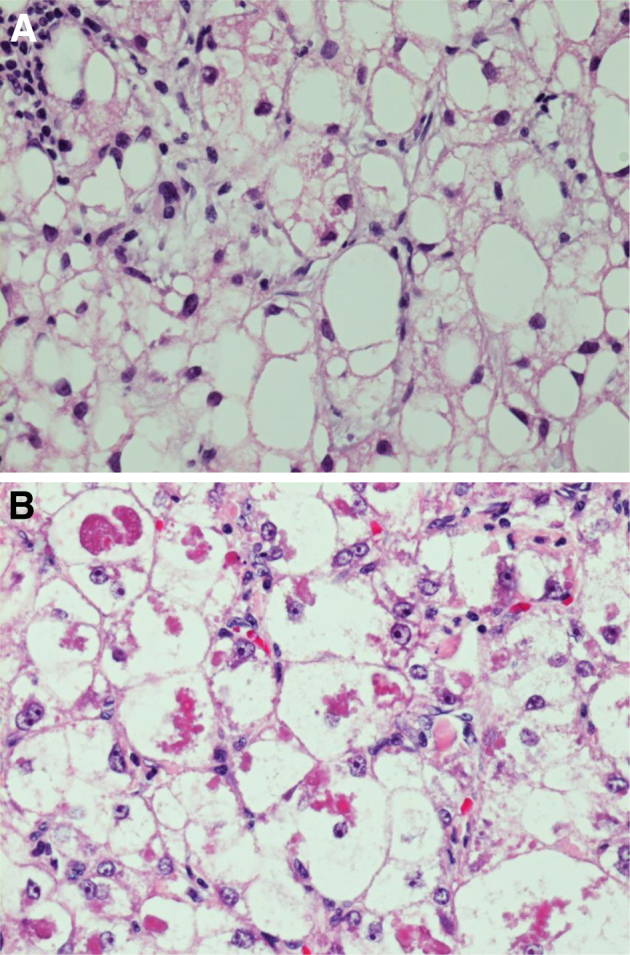
A: H&E staining for hepatocellular carcinoma arising in NASH with prominent intratumoral steatosis, chronic inflammation, and rare ballooned tumor cells containing Mallory-Denk bodies. B: Hepatocellular carcinoma arising in NASH with ballooned tumor cells containing prominent Mallory-Denk bodies. Original magnification, ×40 (A and B).
Patients with NASH-related HCC often have less severe liver dysfunction at HCC diagnosis and may have better overall survival after curative treatments than their counterparts with HCV and/or alcohol-related cirrhosis.42 However, studies specifically evaluating steatohepatitic HCC have not found any difference in prognosis compared to conventional HCC, including similar rates of distant metastases, local recurrence, or overall survival.40
Although there is histological variation between HCC cases, these variations are not routinely used in treatment decision-making. In general, treatment decisions are based on the Barcelona Clinical Liver Cancer staging system, which has been validated in several large cohort studies.43,44 In essence, this staging system incorporates information related to liver function (Child-Turcotte-Pugh Score), performance status, and tumor burden. This staging system is unique in that it has been linked to a treatment algorithm and can be used to triage patients to potentially curative treatments (ie, resection, transplant, ablation) versus chemoembolization versus systemic therapies. It is clear that additional factors could further enhance its prognostic ability, which has spurred the investigation of HCC molecular features that could better portend prognosis or define new opportunities for therapy.
Genetic Underpinnings of HCC
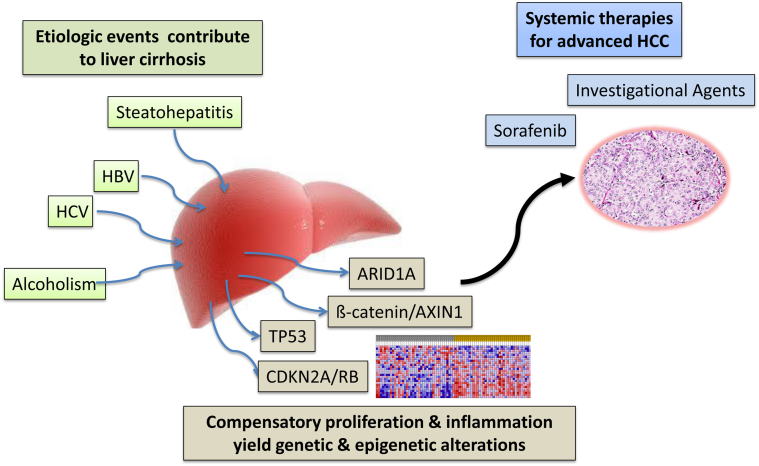
It is clear that diverse etiological stresses play a key role in the etiology of HCC. However, the prevailing view is that liver damage drives aberrant cycles of proliferation in the context of inflamed cirrhotic liver tissue leading to the accumulation of genetic changes that ultimately drive transformation of target cells to HCC (Figure 3).45 Although conventional targeted analyses of specific markers in the HCC have been performed and have identified key pathways of significance to the etiology,46,47 unbiased molecular approaches have provided an opportunity to decipher key pathways and genetic events in disease and their connection to the underlying etiologies. Particularly, utilization of gene expression profiling and next generation sequencing technologies provide a portrait of common molecular events that occur in the pathogenesis of HCC.
Figure 3.
Multiple etiological events drive liver cirrhosis. This results in multiple cycles of compensatory proliferation and inflammation, which can drive the development of genetic and epigenetic alterations. Complex combinations of these events are associated with the development of HCC. Early stage disease can be treated with locoregional therapies, but systemic approaches are relatively limited at present. Multiple investigational drugs are under investigation. HBV, hepatitis B virus; HCV, hepatitis C virus.
At present, the sequencing of exons in approximately 100 HCC cases has been performed (Table 2).48–52 It should be noted that although an important effort from multiple groups, this pales in reference to other diseases, wherein >1000 tumors have been sequenced. However, these studies have yielded a wealth of information in reference to genetic architecture of HCC and key pathways that are likely relevant to the development of HCC.
Table 2.
Studies with Exon Sequencing of HCC Cases
One of the seminal advantages of next generation sequencing is the ability to define new mediators of hepatocarcinogenesis, and the analyses of data have revealed several key previously unknown features of disease and important new pathways that are likely of high relevance in HCC. An important and often underappreciated aspect of sequencing is the ability to define mutation spectra (eg, C to T transitions) that are particularly prevalent in the disease. Many tumor types have distinct spectra that are associated with disease etiology. For example, lung cancers have C→A, which are specifically associated with smoking, whereas melanoma has CC→TT dinucleotide substitutions that would be associated with exposure to solar radiation.53–55 To date, sequencing has been performed largely on cases with a viral (HBV or HCV) or alcohol-related etiology.48–52 In this context, HCC has a relatively unique mutation spectra, G→T, which is apparently modified by the etiological basis of disease, with HCC arising in cirrhotic livers exhibiting differential mutational events relative to noncirrhotic livers.48 However, with the relatively small number of samples, it is difficult to discern if different etiological stresses are associated with specific genetic events. Additionally, to date, no significant cohorts with aflatoxin B1- or NASH-associated tumors have been sequenced; therefore, whether the different mechanisms associated with HCC development will yield distinct genetic alterations remains unknown.
As one may expect, sequencing should identify expected oncogenes and tumor suppressors. The most commonly defined mutated oncogenic driver of HCC is the β-catenin gene (CTNNB1).48–51 This is a validated oncogene that has been previously implicated in HCC pathogenesis from targeted analyses of human tissue and the use of mouse models.56 Further implicating the importance of this pathway, loss of Axin1 and Axin2, which are negative regulators of β-catenin, are also frequently observed in HCC.48–51 Similarly, TP53 has been identified as a frequently mutated tumor suppressor in HCC.48–51 Again, this is consistent with established analysis of human tumor specimens. Surprisingly, TP53 itself is a weak tumor suppressor in mouse models, and largely cooperates with other stresses in suppressing the development of HCC or curtailing the metastatic potential of HCC models.57,58 These findings reinforce the importance of investigating human tissue to truly understand the underpinnings of human disease. Because exome or whole genome sequencing is a discovery platform, it was an important finding that multiple chromatin modifiers of the SWI/SNF family are associated with HCC.48–51 Loss of these tumor suppressors is relatively common across multiple solid tumors.59 Importantly, in such analyses, although Arid1A and Arid2 mutations are particularly prevalent in HCC, multiple genes in the SWI/SNF chromatin remodeling complex are mutated. However, the functional significance of such alterations remains obscure relative to the etiology or progression of HCC. A large number of additional genes involved in discrete carcinogenic processes involved in carcinogenesis were found to be mutated in HCC. For example, composite mutations/loss of cell cycle regulatory genes CDKN2A and RB1, disruption of genes involved in chromosome stability such as ATM and MLH1, and genes that mediate epigenetic regulation, such as MLL3 and MLL, have all been found to be mutated in HCC.48–51 Although these data yield some insight into the genetics of HCC, the lack of commonality between individual cancer cases is most striking. This contrasts with tumors like pancreatic cancer in which veritably of all cases harbor well-known oncogenic drivers (ie, K-ras). These data underline one of the unique features of HCC in that it is highly heterogeneous. Given this same heterogeneity it remains unclear how such genetic findings will yield actionable insights on which disease treatment can be modified to improve patient outcomes.
Gene Expression Profiling Prognostic Markers and Pathways
In contrast with the seemingly infinite diversity of HCC as shown by sequencing, gene expression profiling approaches monitoring changes in RNA levels on microarray platforms have defined a limited series of HCC subtypes (Table 3).
Table 3.
HCC Subtypes Identified by Gene Expression Profiling Approaches
| Subtypes | Genes/Processes | Clinical endpoint | References |
|---|---|---|---|
| A and B | Multiple/proliferation | Overall survival | 61 |
| CIN: high and low | Proliferation | Overall survival | 64 |
| G1-G6 | Multiple/proliferation | Overall survival | 67 |
| RB-loss: high and low | Proliferation | Overall survival | 63 |
| A, B, and C | Multiple/proliferation | Overall survival | 65 |
| 65 genes | Multiple | Overall survival | 72 |
| 5-gene score | HN1, RAN, RAMP3, KRT19, and TAF9 | Overall survival End-stage disease |
70 |
| 186-gene signature | Nontumor liver | Overall survival Risk of recurrence |
69 |
CIN, chromosome instability signature.
Early work from Dr. Thorgeirrson's group defined gene expression changes in HCC cases related to proliferation and other core features of disease that were associated with prognosis.60–63 These findings were recapitulated from numerous groups demonstrating that tumors with elevated expression of proliferation-associated genes were prognostic for poor outcome dependent on methodology.34,63–66 Additional analyses served to define subgroups. Specifically, HCC can be classified into three or six subtypes based on gene expression profiling.65,67 These groups are consistent across multiple independent cohorts and have differential overall prognosis.65 This work was similar to that performed in breast cancer68 and is important for clarifying intrinsically different forms of HCC that have distinct prognosis. Such multigene signatures have been distilled to inform progression of early stage disease and vascular invasion.31,69–72 Most recently, extensive analysis of clinical specimens by gene expression profiling defined a 5-gene signature.70 This signature was effective in both training and validation sets and outperformed other signatures that have been previously identified for determining the risk of recurrence and death after disease resection. Analysis of the nontumor liver tissue also provides important prognostic information. A 186-gene signature is associated with poor prognosis of early stage disease and risk of disease progression in the cirrhotic liver.69 Presumably, this is because the tissue is reflective of the carcinogenic milieu within the liver, and thus it is associated with risk of poor clinical outcome. Although multiple signatures appear prognostic in retrospective analysis, there remain no established predictive markers that have been shown to improve patient treatment. Thus, although there are newly defined subtypes of HCC based on gene expression profiles, these markers have yet to yield directed treatments for HCC.
Targeted Therapeutic Interventions: A Work in Progress
Despite substantial and accelerated research in the area of HCC, the 5-year survival for advanced disease remains <10%.73–75 Localized disease can be treated with ablative therapies, surgical resection, or transplantion.75–77 However, many cases present at the advanced stage or the patient is a poor candidate for such interventions. Thus, substantial clinical research has been conducted to provide improved treatment for HCC with systemic therapies. Phase III trials have evaluated multiple therapeutic modalities, including tamoxifen, chemotherapy, and interferon, all with negative outcomes.76,78,79 The lack of benefit with multiple chemotherapy regimens makes HCC somewhat unique in cancer by having no standard cytotoxic therapy, and indicates that HCC must be treated by other targeted agents.80–82 The only drug approved for advanced HCC is the multikinase inhibitor sorafenib. In HCC, sorafenib functions as a cytostatic agent that provides an approximate 3-month improvement in survival versus placebo control based on large randomized phase III trial versus placebo.83 Despite the positive impact on survival, HCC remains a classical therapy recalcitrant disease for which new interventions are needed.
Multiple ongoing phase II and phase III trials support the possible development of new treatment opportunities for advanced HCC.78,84 The majority of phase II trials represent approaches to define positive effect of treatment with progression-free survival as the primary endpoint.85,86 These studies build off of the clinical outcomes with sorafenib and are often conducted in the second line (after sorafenib failure) or as a first-line agent in combination with sorafenib.84 A number of agents have shown promise in this context including brivanib and bevacizumab-erlotinib.87–90 Because of the advanced nature of HCC and the underlying liver disease and underlying liver disease associated with late stage HCC, the outcomes have been relatively modest. Interestingly, sorafenib, brivanib, and bevacizumab all targeted VEGF signaling, suggesting that this may be an important therapeutic node in HCC. Based on the identification of multiple additional key pathways disrupted in HCC, trials have been rationally directed to target common oncogenic events. For example, with the key role of Met signaling in HCC, the drug tivantinib has been investigated in a randomized phase II trial.91,92 Similarly, given the frequent loss of CDKN2A and deregulation of the retinoblastoma tumor suppressor pathway, the CDK4/6 inhibitor PD-0332991 is being tested in advanced HCC (http://clinicaltrials.gov, Clinical Trial NCT01356628). These trials and further multiple, additional trials will likely shed light on agents that have potential usefulness in HCC. A critical aspect of such targeted therapies is the importance of defining features of disease that underlie sensitivity to the agent. For example, in retrospective analysis, tivantinib is particularly effective against HCC, which has a high expression of Met.91,92 Therefore, it will be important to leverage the molecular analyses of HCC to delineate specific subtypes of HCC or individual cases that would be particularly responsive to a given therapeutic regimen.
Challenges and Future Directions
HCC represents a particular challenge for which there is multiple opportunities to change the large burden of disease and terrible prognosis.
Prevention
Because etiological features of HCC are very well-defined, it represents a disease for which preventive strategies could be particularly effective. Ostensibly, these etiologies can be ultimately controlled, thereby substantially reducing the number of individuals at high risk for HCC. The decreasing incidence of HCC in Asia speaks to the potential benefits and promise of preventive measures, such as HBV vaccination and treatment.93 Although a vaccine for HCV does not exist, successful treatment has been associated with significant reductions in HCC risk.94 The introduction of interferon-free treatment regimens, with better success rates and lower toxicity rates, may further expand HCV treatment rates and help to reduce the future burden of HCC.95 Given the increasing prevalence of NASH-related HCC, studies assessing the potential impact of weight loss, either via diet and exercise or bariatric surgery, are necessary. Finally, larger studies assessing the potential preventive benefit of diet, coffee, and statins are necessary.96,97
Early Detection
Because the etiological factors of HCC can be defined relatively easily in the clinic, it is surprising that the diagnosis of HCC often occurs at such a late stage.18 This failure is related to several reasons, including underutilization of surveillance, delayed follow-up of surveillance tests, and suboptimal effectiveness of surveillance tests. Several studies have demonstrated that <20% of patients with cirrhosis receive HCC surveillance, as recommended by guidelines.98–100 Underutilization of HCC surveillance is related to multiple failure points, including under-recognition of liver disease, under-recognition of cirrhosis, and physicians failing to order HCC surveillance in patients with known cirrhosis, highlighting a need for multifaceted interventions to improve cirrhosis recognition and facilitate HCC surveillance testing.101 However, a secondary analysis of the Hepatitis C Antiviral Long-term Treatment Against Cirrhosis (HALT-C) trial suggested that the most common reason for late stage tumor presentation is related to poor sensitivity of currently available surveillance tools.102 Therefore, it is critically important to develop markers that are easily used that have a high sensitivity and specificity for the detection of early stage disease. Such markers could then provide the trigger for radiological imaging to identify early stage disease in which resection or ablative therapies can be more effective.
Fully Understanding the Genetic Diversity
For HCC, sequencing analysis has served to provide a better understanding of the tumor genetics. However, these data have also demonstrated that no HCC tumor is alike, and only a minority of cases has a mutation that would immediately provoke a specific therapeutic approach. Therefore, the challenge is to understand how genetics can be used to provide a road map for the treatment of HCC.
Dissecting the Therapeutic Sensitivities of HCC Subtypes
At present, HCC cases are treated similarly with little consideration of molecular subtypes or markers that could be relevant to prognosis or therapeutic sensitivities. There is clearly an opportunity to define markers predictive of response to specific agents to begin refining the treatment paradigm for HCC from a one size fits all to one of more precision-guided therapy. The emerging trials with tivantinib will provide the first indication that stratifying patients to specific treatment regimens will perhaps improve care.91
Implementing More Effective Treatment
The dismal prognosis of advanced HCC provides a seminal opportunity for improved treatment options. The plethora of ongoing early stage trials provides hope for more effective regimens, although it will be critically important to consider the incredible diversity of HCC when investigating therapeutic response. Clinicians should be cognizant of the opportunity that is provided by analyzing tissue from such trials to better inform future clinical interventions and to avoid making broad conclusions on an unselected patient population, wherein the expectation should be likely that only a subset of patients would experience a favorable response to the intervention.
Footnotes
Supported by grants from NIH (E.S.K.) and the Melanoma Research Association (E.S.K.) and by contract funding from Pfizer (E.S.K.).
Disclosures: A.S. is on the Speaker Bureau for Onyx Pharmaceuticals. E.S.K. is a consultant for Pfizer.
Contributor Information
Erik S. Knudsen, Email: erik.knudsen@utsouthwestern.edu.
Amit G. Singal, Email: amit.singal@utsouthwestern.edu.
References
- 1.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley R. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 4.Yu M.W., Chang H.C., Liaw Y.F., Lin S.M., Lee S.D., Liu C.J., Chen P.J., Hsiao T.J., Lee P.H., Chen C.J. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst. 2000;92:1159–1164. doi: 10.1093/jnci/92.14.1159. [DOI] [PubMed] [Google Scholar]
- 5.Kew M.C., Macerollo P. Effect of age on the etiologic role of the hepatitis B virus in hepatocellular carcinoma in blacks. Gastroenterology. 1988;94:439–442. doi: 10.1016/0016-5085(88)90434-9. [DOI] [PubMed] [Google Scholar]
- 6.Qian G.S., Ross R.K., Yu M.C., Yuan J.M., Gao Y.T., Henderson B.E., Wogan G.N., Groopman J.D. A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People's Republic of China. Cancer Epidemiol Biomarkers Prev. 1994;3:3–10. [PubMed] [Google Scholar]
- 7.El-Serag H.B. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Donato F., Boffetta P., Puoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int J Cancer. 1998;75:347–354. doi: 10.1002/(sici)1097-0215(19980130)75:3<347::aid-ijc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Fattovich G., Giustina G., Degos F., Tremolada F., Diodati G., Almasio P., Nevens F., Solinas A., Mura D., Brouwer J.T., Thomas H., Njapoum C., Casarin C., Bonetti P., Fuschi P., Basho J., Tocco A., Bhalla A., Galassini R., Noventa F., Schalm S.W., Realdi G. Morbidity and mortality in compensated cirrhosis type C: a retrospective follow-up study of 384 patients. Gastroenterology. 1997;112:463–472. doi: 10.1053/gast.1997.v112.pm9024300. [DOI] [PubMed] [Google Scholar]
- 10.Sun C.A., Wu D.M., Lin C.C., Lu S.N., You S.L., Wang L.Y., Wu M.H., Chen C.J. Incidence and cofactors of hepatitis C virus-related hepatocellular carcinoma: a prospective study of 12,008 men in Taiwan. Am J Epidemiol. 2003;157:674–682. doi: 10.1093/aje/kwg041. [DOI] [PubMed] [Google Scholar]
- 11.Adami H.O., Hsing A.W., McLaughlin J.K., Trichopoulos D., Hacker D., Ekbom A., Persson I. Alcoholism and liver cirrhosis in the etiology of primary liver cancer. Int J Cancer. 1992;51:898–902. doi: 10.1002/ijc.2910510611. [DOI] [PubMed] [Google Scholar]
- 12.Hassan M.M., Hwang L.Y., Hatten C.J., Swaim M., Li D., Abbruzzese J.L., Beasley P., Patt Y.Z. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206–1213. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- 13.Starley B.Q., Calcagno C.J., Harrison S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 14.White D.L., Kanwal F., El-Serag H.B. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10:1342–1359.e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tajiri H., Tanaka H., Brooks S., Takano T. Reduction of hepatocellular carcinoma in childhood after introduction of selective vaccination against hepatitis B virus for infants born to HBV carrier mothers. Cancer Causes Control. 2011;22:523–527. doi: 10.1007/s10552-010-9721-4. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y., Hanada K., Mizokami M., Yeo A.E., Shih J.W., Gojobori T., Alter H.J. A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc Natl Acad Sci U S A. 2002;99:15584–15589. doi: 10.1073/pnas.242608099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umemura T., Ichijo T., Yoshizawa K., Tanaka E., Kiyosawa K. Epidemiology of hepatocellular carcinoma in Japan. J Gastroenterol. 2009;44(Suppl 19):102–107. doi: 10.1007/s00535-008-2251-0. [DOI] [PubMed] [Google Scholar]
- 18.Altekruse S.F., McGlynn K.A., Reichman M.E. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singal A.G., Mukherjee A., Elmunzer B.J., Higgins P.D.R., Lok A.S., Zhu J., Marrero J.A., Waljee A.K. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108:1723–1730. doi: 10.1038/ajg.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen M.F., Tanaka Y., Fong D.Y., Fung J., Wong D.K., Yuen J.C., But D.Y., Chan A.O., Wong B.C., Mizokami M., Lai C.L. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol. 2009;50:80–88. doi: 10.1016/j.jhep.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Chalasani N., Said A., Ness R., Hoen H., Lumeng L. Screening for hepatocellular carcinoma in patients with cirrhosis in the United States: results of a national survey. Am J Gastroenterol. 1999;94:2224–2229. doi: 10.1111/j.1572-0241.1999.01297.x. [DOI] [PubMed] [Google Scholar]
- 22.Singal A., Volk M.L., Waljee A., Salgia R., Higgins P., Rogers M.A., Marrero J.A. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther. 2009;30:37–47. doi: 10.1111/j.1365-2036.2009.04014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruix J., Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2010;53:1–35. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee E., Edward S., Singal A.G., Lavieri M.S., Volk M. Improving screening for hepatocellular carcinoma by incorporating data on levels of alpha-fetoprotein, over time. Clin Gastroenterol Hepatol. 2013;11:437–440. doi: 10.1016/j.cgh.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 25.Marrero J.A., El-Serag H.B. Alpha-fetoprotein should be included in the hepatocellular carcinoma surveillance guidelines of the American Association for the Study of Liver Diseases. Hepatology. 2011;53:1060–1061. doi: 10.1002/hep.24033. [DOI] [PubMed] [Google Scholar]
- 26.Singal A.G., Conjeevaram H.S., Volk M.L., Fu S., Fontana R.J., Askari F., Su G.L., Lok A.S., Marrero J.A. Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis. Cancer Epidemiol Biomarkers Prev. 2012;21:793–799. doi: 10.1158/1055-9965.EPI-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopal P., Yopp A.C., Waljee A.K., Chiang J., Nehra M., Kandunoori P., Singal A.G. Factors that affect accuracy of alpha-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. Clin Gastroenterol Hepatol. 2013 doi: 10.1016/j.cgh.2013.09.053. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato Y., Nakata K., Kato Y., Shima M., Ishii N., Koji T., Taketa K., Endo Y., Nagataki S. Early recognition of hepatocellular carcinoma based on altered profiles of alpha-fetoprotein. N Engl J Med. 1993;328:1802–1806. doi: 10.1056/NEJM199306243282502. [DOI] [PubMed] [Google Scholar]
- 29.Marrero J.A., Feng Z., Wang Y., Nguyen M.H., Befeler A.S., Roberts L.R., Reddy K.R., Harnois D., Llovet J.M., Normolle D., Dalhgren J., Chia D., Lok A.S., Wagner P.D., Srivastava S., Schwartz M. Alpha-fetoprotein, Des-gamma Carboxyprothrombin, and Lectin-Bound Alpha-fetoprotein in Early Hepatocellular Carcinoma AFP, DCP, and AFP-L3 in Hepatocellular carcinoma. Gastroenterology. 2009;137:110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capurro M., Wanless I.R., Sherman M., Deboer G., Shi W., Miyoshi E., Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 31.Llovet J.M., Chen Y., Wurmbach E., Roayaie S., Fiel M.I., Schwartz M., Thung S.N., Khitrov G., Zhang W., Villanueva A., Battiston C., Mazzaferro V., Bruix J., Waxman S., Friedman S.L. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Marrero J.A., Romano P.R., Nikolaeva O., Steel L., Mehta A., Fimmel C.J., Comunale M.A., D'Amelio A., Lok A.S., Block T.M. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Shang S., Plymoth A., Ge S., Feng Z., Rosen H.R., Sangrajrang S., Hainaut P., Marrero J.A., Beretta L. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 2012;55:483–490. doi: 10.1002/hep.24703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beneduce L., Castaldi F., Marino M., Quarta S., Ruvoletto M., Benvegnu L., Calabrese F., Gatta A., Pontisso P., Fassina G. Squamous cell carcinoma antigen-immunoglobulin M complexes as novel biomarkers for hepatocellular carcinoma. Cancer. 2005;103:2558–2565. doi: 10.1002/cncr.21106. [DOI] [PubMed] [Google Scholar]
- 35.Yamagami H., Moriyama M., Tanaka N., Arakawa Y. Detection of serum and intrahepatic human hepatocyte growth factor in patients with type C liver diseases. Intervirology. 2001;44:36–42. doi: 10.1159/000050028. [DOI] [PubMed] [Google Scholar]
- 36.Mazziotti G., Sorvillo F., Morisco F., Carbone A., Rotondi M., Stornaiuolo G., Precone D.F., Cioffi M., Gaeta G.B., Caporaso N., Carella C. Serum insulin-like growth factor I evaluation as a useful tool for predicting the risk of developing hepatocellular carcinoma in patients with hepatitis C virus-related cirrhosis: a prospective study. Cancer. 2002;95:2539–2545. doi: 10.1002/cncr.11002. [DOI] [PubMed] [Google Scholar]
- 37.Chignard N., Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127:S120–S125. doi: 10.1053/j.gastro.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Alexander J., Torbenson M., Wu T.T., Yeh M.M. Non-alcoholic fatty liver disease contributes to hepatocarcinogenesis in non-cirrhotic liver: a clinical and pathological study. J Gastroenterol Hepatol. 2013;28:848–854. doi: 10.1111/jgh.12116. [DOI] [PubMed] [Google Scholar]
- 39.Salomao M., Yu W.M., Brown R.S., Jr., Emond J.C., Lefkowitch J.H. Steatohepatitic hepatocellular carcinoma (SH-HCC): a distinctive histological variant of HCC in hepatitis C virus-related cirrhosis with associated NAFLD/NASH. Am J Surg Pathol. 2010;34:1630–1636. doi: 10.1097/PAS.0b013e3181f31caa. [DOI] [PubMed] [Google Scholar]
- 40.Salomao M., Remotti H., Vaughan R., Siegel A.B., Lefkowitch J.H., Moreira R.K. The steatohepatitic variant of hepatocellular carcinoma and its association with underlying steatohepatitis. Hum Pathol. 2012;43:737–746. doi: 10.1016/j.humpath.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Jain D., Nayak N.C., Kumaran V., Saigal S. Steatohepatitic hepatocellular carcinoma, a morphologic indicator of associated metabolic risk factors: a study from India. Arch Pathol Lab Med. 2013;137:961–966. doi: 10.5858/arpa.2012-0048-OA. [DOI] [PubMed] [Google Scholar]
- 42.Reddy S.K., Steel J.L., Chen H.W., DeMateo D.J., Cardinal J., Behari J., Humar A., Marsh J.W., Geller D.A., Tsung A. Outcomes of curative treatment for hepatocellular cancer in nonalcoholic steatohepatitis versus hepatitis C and alcoholic liver disease. Hepatology. 2012;55:1809–1819. doi: 10.1002/hep.25536. [DOI] [PubMed] [Google Scholar]
- 43.Marrero J.A., Fontana R.J., Barrat A., Askari F., Conjeevaram H.S., Su G.L., Lok A.S. Prognosis of hepatocellular carcinoma: comparison of 7 staging systems in an American cohort. Hepatology. 2005;41:707–716. doi: 10.1002/hep.20636. [DOI] [PubMed] [Google Scholar]
- 44.Cillo U., Vitale A., Grigoletto F., Farinati F., Brolese A., Zanus G., Neri D., Boccagni P., Srsen N., D'Amico F., Ciarleglio F.A., Bridda A., D'Amico D.F. Prospective validation of the Barcelona Clinic Liver Cancer staging system. J Hepatol. 2006;44:723–731. doi: 10.1016/j.jhep.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 46.Buendia M.A. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 47.Bioulac-Sage P., Laurent-Puig P., Balabaud C., Zucman-Rossi J. Genetic alterations in hepatocellular adenomas. Hepatology. 2003;37:480. doi: 10.1053/jhep.2003.50058. author reply -1. [DOI] [PubMed] [Google Scholar]
- 48.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., Calderaro J., Bioulac-Sage P., Letexier M., Degos F., Clement B., Balabaud C., Chevet E., Laurent A., Couchy G., Letouze E., Calvo F., Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang J., Deng Q., Wang Q., Li K.Y., Dai J.H., Li N., Zhu Z.D., Zhou B., Liu X.Y., Liu R.F., Fei Q.L., Chen H., Cai B., Zhou B., Xiao H.S., Qin L.X., Han Z.G. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 50.Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 51.Kan Z., Zheng H., Liu X., Li S., Barber T.D., Gong Z. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa H., Shibata T. Comprehensive genome sequencing of the liver cancer genome. Cancer Lett. 2013;340:234–240. doi: 10.1016/j.canlet.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 53.Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krauthammer M., Kong Y., Ha B.H., Evans P., Bacchiocchi A., McCusker J.P., Cheng E., Davis M.J., Goh G., Choi M., Ariyan S., Narayan D., Dutton-Regester K., Capatana A., Holman E.C., Bosenberg M., Sznol M., Kluger H.M., Brash D.E., Stern D.F., Materin M.A., Lo R.S., Mane S., Ma S., Kidd K.K., Hayward N.K., Lifton R.P., Schlessinger J., Boggon T.J., Halaban R. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikolaev S.I., Rimoldi D., Iseli C., Valsesia A., Robyr D., Gehrig C., Harshman K., Guipponi M., Bukach O., Zoete V., Michielin O., Muehlethaler K., Speiser D., Beckmann J.S., Xenarios I., Halazonetis T.D., Jongeneel C.V., Stevenson B.J., Antonarakis S.E. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 56.Harada N., Oshima H., Katoh M., Tamai Y., Oshima M., Taketo M.M. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 2004;64:48–54. doi: 10.1158/0008-5472.can-03-2123. [DOI] [PubMed] [Google Scholar]
- 57.McClendon A.K., Dean J.L., Ertel A., Fu Z., Rivadeneira D.B., Reed C.A., Bourgo R.J., Witkiewicz A., Addya S., Mayhew C.N., Grimes H.L., Fortina P., Knudsen E.S. RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology. 2011;141:1439–1450. doi: 10.1053/j.gastro.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis B.C., Klimstra D.S., Socci N.D., Xu S., Koutcher J.A., Varmus H.E. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol Cell Biol. 2005;25:1228–1237. doi: 10.1128/MCB.25.4.1228-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J.S., Thorgeirsson S.S. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134–1143. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
- 61.Lee J.S., Chu I.S., Heo J., Calvisi D.F., Sun Z., Roskams T., Durnez A., Demetris A.J., Thorgeirsson S.S. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 62.Lee J.S., Thorgeirsson S.S. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 63.Mayhew C.N., Carter S.L., Fox S.R., Sexton C.R., Reed C.A., Srinivasan S.V., Liu X., Wikenheiser-Brokamp K., Boivin G.P., Lee J.S., Aronow B.J., Thorgeirsson S.S., Knudsen E.S. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 64.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 65.Hoshida Y., Nijman S.M., Kobayashi M., Chan J.A., Brunet J.P., Chiang D.Y., Villanueva A., Newell P., Ikeda K., Hashimoto M., Watanabe G., Gabriel S., Friedman S.L., Kumada H., Llovet J.M., Golub T.R. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villanueva A., Hoshida Y., Battiston C., Tovar V., Sia D., Alsinet C., Cornella H., Liberzon A., Kobayashi M., Kumada H., Thung S.N., Bruix J., Newell P., April C., Fan J.B., Roayaie S., Mazzaferro V., Schwartz M.E., Llovet J.M. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512.e2. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyault S., Rickman D.S., de Reynies A., Balabaud C., Rebouissou S., Jeannot E., Herault A., Saric J., Belghiti J., Franco D., Bioulac-Sage P., Laurent-Puig P., Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 68.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lonning P.E., Borresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 69.Hoshida Y., Villanueva A., Sangiovanni A., Sole M., Hur C., Andersson K.L., Chung R.T., Gould J., Kojima K., Gupta S., Taylor B., Crenshaw A., Gabriel S., Minguez B., Iavarone M., Friedman S.L., Colombo M., Llovet J.M., Golub T.R. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nault J.C., De Reynies A., Villanueva A., Calderaro J., Rebouissou S., Couchy G., Decaens T., Franco D., Imbeaud S., Rousseau F., Azoulay D., Saric J., Blanc J.F., Balabaud C., Bioulac-Sage P., Laurent A., Laurent-Puig P., Llovet J.M., Zucman-Rossi J. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 71.Minguez B., Hoshida Y., Villanueva A., Toffanin S., Cabellos L., Thung S., Mandeli J., Sia D., April C., Fan J.B., Lachenmayer A., Savic R., Roayaie S., Mazzaferro V., Bruix J., Schwartz M., Friedman S.L., Llovet J.M. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J Hepatol. 2011;55:1325–1331. doi: 10.1016/j.jhep.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim S.M., Leem S.H., Chu I.S., Park Y.Y., Kim S.C., Kim S.B., Park E.S., Lim J.Y., Heo J., Kim Y.J., Kim D.G., Kaseb A., Park Y.N., Wang X.W., Thorgeirsson S.S., Lee J.S. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bruix J., Llovet J.M. Major achievements in hepatocellular carcinoma. Lancet. 2009;373:614–616. doi: 10.1016/S0140-6736(09)60381-0. [DOI] [PubMed] [Google Scholar]
- 74.Olsen S.K., Brown R.S., Siegel A.B. Hepatocellular carcinoma: review of current treatment with a focus on targeted molecular therapies. Therap Adv Gastroenterol. 2010;3:55–66. doi: 10.1177/1756283X09346669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 76.de Lope C.R., Tremosini S., Forner A., Reig M., Bruix J. Management of HCC. J Hepatol. 2012;56(Suppl 1):S75–S87. doi: 10.1016/S0168-8278(12)60009-9. [DOI] [PubMed] [Google Scholar]
- 77.Rossi L., Zoratto F., Papa A., Iodice F., Minozzi M., Frati L., Tomao S. Current approach in the treatment of hepatocellular carcinoma. World J Gastrointest Oncol. 2010;2:348–359. doi: 10.4251/wjgo.v2.i9.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chua C.W., Choo S.P. Targeted therapy in hepatocellular carcinoma. Int J Hepatol. 2011;2011:348297. doi: 10.4061/2011/348297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Worns M.A., Weinmann A., Schuchmann M., Galle P.R. Systemic therapies in hepatocellular carcinoma. Dig Dis. 2009;27:175–188. doi: 10.1159/000218351. [DOI] [PubMed] [Google Scholar]
- 80.Llovet J.M., Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Llovet J.M., Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008. J Hepatol. 2008;48(Suppl 1):S20–S37. doi: 10.1016/j.jhep.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 82.Finn R.S. Development of molecularly targeted therapies in hepatocellular carcinoma: where do we go now? Clin Cancer Res. 2010;16:390–397. doi: 10.1158/1078-0432.CCR-09-2084. [DOI] [PubMed] [Google Scholar]
- 83.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T.F., Galle P.R., Seitz J.F., Borbath I., Haussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., Group S.I.S. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 84.Thomas M.B., Jaffe D., Choti M.M., Belghiti J., Curley S., Fong Y., Gores G., Kerlan R., Merle P., O'Neil B., Poon R., Schwartz L., Tepper J., Yao F., Haller D., Mooney M., Venook A. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28:3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Llovet J.M., Di Bisceglie A.M., Bruix J., Kramer B.S., Lencioni R., Zhu A.X., Sherman M., Schwartz M., Lotze M., Talwalkar J., Gores G.J. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 86.Llovet J.M., Bruix J. Testing molecular therapies in hepatocellular carcinoma: the need for randomized phase II trials. J Clin Oncol. 2009;27:833–835. doi: 10.1200/JCO.2008.19.1973. [DOI] [PubMed] [Google Scholar]
- 87.Finn R.S., Kang Y.K., Mulcahy M., Polite B.N., Lim H.Y., Walters I., Baudelet C., Manekas D., Park J.W. Phase II, open-label study of brivanib as second-line therapy in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2012;18:2090–2098. doi: 10.1158/1078-0432.CCR-11-1991. [DOI] [PubMed] [Google Scholar]
- 88.Park J.W., Finn R.S., Kim J.S., Karwal M., Li R.K., Ismail F., Thomas M., Harris R., Baudelet C., Walters I., Raoul J.L. Phase II, open-label study of brivanib as first-line therapy in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2011;17:1973–1983. doi: 10.1158/1078-0432.CCR-10-2011. [DOI] [PubMed] [Google Scholar]
- 89.Thomas M.B., Chadha R., Glover K., Wang X., Morris J., Brown T., Rashid A., Dancey J., Abbruzzese J.L. Phase 2 study of erlotinib in patients with unresectable hepatocellular carcinoma. Cancer. 2007;110:1059–1067. doi: 10.1002/cncr.22886. [DOI] [PubMed] [Google Scholar]
- 90.Thomas M.B., Morris J.S., Chadha R., Iwasaki M., Kaur H., Lin E., Kaseb A., Glover K., Davila M., Abbruzzese J. Phase II trial of the combination of bevacizumab and erlotinib in patients who have advanced hepatocellular carcinoma. J Clin Oncol. 2009;27:843–850. doi: 10.1200/JCO.2008.18.3301. [DOI] [PubMed] [Google Scholar]
- 91.Santoro A., Rimassa L., Borbath I., Daniele B., Salvagni S., Van Laethem J.L., Van Vlierberghe H., Trojan J., Kolligs F.T., Weiss A., Miles S., Gasbarrini A., Lencioni M., Cicalese L., Sherman M., Gridelli C., Buggisch P., Gerken G., Schmid R.M., Boni C., Personeni N., Hassoun Z., Abbadessa G., Schwartz B., Von Roemeling R., Lamar M.E., Chen Y., Porta C. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol. 2013;14:55–63. doi: 10.1016/S1470-2045(12)70490-4. [DOI] [PubMed] [Google Scholar]
- 92.Trojan J., Zeuzem S. Tivantinib in hepatocellular carcinoma. Expert Opin Investig Drugs. 2013;22:141–147. doi: 10.1517/13543784.2013.741586. [DOI] [PubMed] [Google Scholar]
- 93.Chang M.H., Chen C.J., Lai M.S., Hsu H.M., Wu T.C., Kong M.S., Liang D.C., Shau W.Y., Chen D.S. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N Engl J Med. 1997;336:1855–1859. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 94.Singal A.G., Volk M.L., Jensen D., Di Bisceglie A.M., Schoenfeld P.S. A sustained viral response is associated with reduced liver-related morbidity and mortality in patients with hepatitis C virus. Clin Gastroenterol Hepatol. 2010;8:280–288. doi: 10.1016/j.cgh.2009.11.018. 288.e1. [DOI] [PubMed] [Google Scholar]
- 95.Lok A.S., Gardiner D.F., Lawitz E., Martorell C., Everson G.T., Ghalib R., Reindollar R., Rustgi V., McPhee F., Wind-Rotolo M., Persson A., Zhu K., Dimitrova D.I., Eley T., Guo T., Grasela D.M., Pasquinelli C. Preliminary study of two antiviral agents for hepatitis C genotype 1. N Engl J Med. 2012;366:216–224. doi: 10.1056/NEJMoa1104430. [DOI] [PubMed] [Google Scholar]
- 96.Bravi F., Bosetti C., Tavani A., La Vecchia C. Coffee drinking and hepatocellular carcinoma: an update. Hepatology. 2009;50:1317–1318. doi: 10.1002/hep.23272. [DOI] [PubMed] [Google Scholar]
- 97.Singh S., Singh P.P., Singh A.G., Murad M.H., Sanchez W. Statins Are Associated With a Reduced Risk of Hepatocellular Cancer: A Systematic Review and Meta-analysis. Gastroenterology. 2013;144:323–332. doi: 10.1053/j.gastro.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 98.Davila J.A., Henderson L., Kramer J.R., Kanwal F., Richardson P.A., Duan Z., El-Serag H.B. Utilization of surveillance for hepatocellular carcinoma among hepatitis C virus-infected veterans in the United States. Ann Intern Med. 2011;154:85–93. doi: 10.7326/0003-4819-154-2-201101180-00006. [DOI] [PubMed] [Google Scholar]
- 99.Singal A.G., Yopp A., SS C., Packer M., Lee W.M., Tiro J.A. Utilization of hepatocellular carcinoma surveillance among American patients: a systematic review. J Gen Intern Med. 2012;27:861–867. doi: 10.1007/s11606-011-1952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singal A.G., Yopp A., Gupta S., Skinner C.S., Halm E.A., Okolo E., Nehra M., Lee W.M., Marrero J.A., Tiro J.A. Failure rates in the hepatocellular carcinoma surveillance process. Cancer Prevent Res. 2012;5:1124–1130. doi: 10.1158/1940-6207.CAPR-12-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singal A.G., Tiro J.A., Gupta S. Improving hepatocellular carcinoma screening: applying lessons from colorectal cancer screening. Clin Gastroenterol Hepatol. 2013;11:472–477. doi: 10.1016/j.cgh.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singal A.G., Nehra M., Adams-Huet B., Yopp A.C., Tiro J.A., Marrero J.A., Lok A.S., Lee W.M. Detection of hepatocellular carcinoma at advanced stages among patients in the HALT-C trial: where did surveillance fail? Am J Gastroenterol. 2013;108:425–432. doi: 10.1038/ajg.2012.449. [DOI] [PMC free article] [PubMed] [Google Scholar]