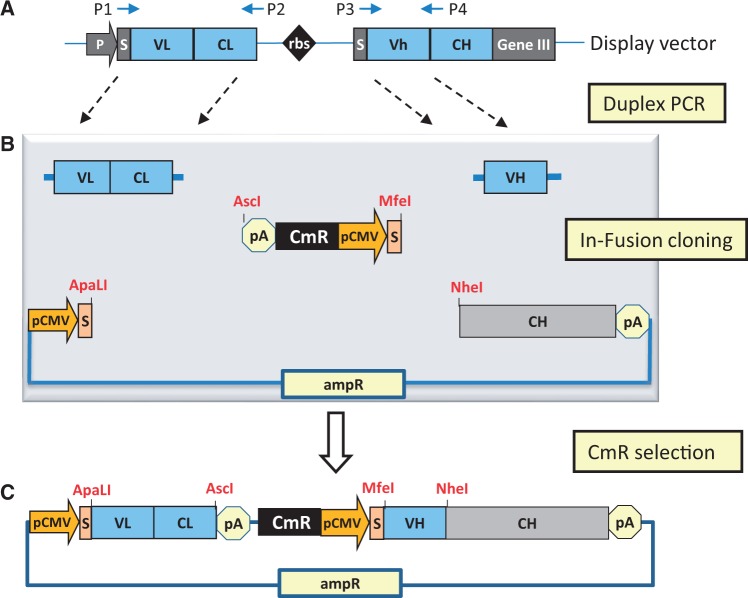
Figure 1.
Schematic representation of zero-background IgG reformatting using InTag positive selection. (A) Amplification of variable antibody regions from phagemid. The light chain and VH-encoding regions are amplified from the phage display construct by duplex-PCR with primers P1, P2, P3 and P4 (Supplementary Table S1). (B) In-Fusion cloning of variable antibody regions using InTag positive selection. PCR products from (A) were treated with cloning enhancer and mixed with pre-prepared InTag adaptor, mammalian expression vector (with heavy chain constant region) and the In-Fusion cloning enzyme. The resulting DNA was transformed into E. coli, and recombinant plasmids were selected in liquid media containing chloramphenicol. P: promoter; S: signal peptide; rbs: ribosome binding site; pA: polyadenylation signal, pCMV: CMV promoter, CmR: chloramphenicol-resistance marker, AmpR: ampicillin resistance marker. (C) Final IgG reformatted mammalian expression vector.