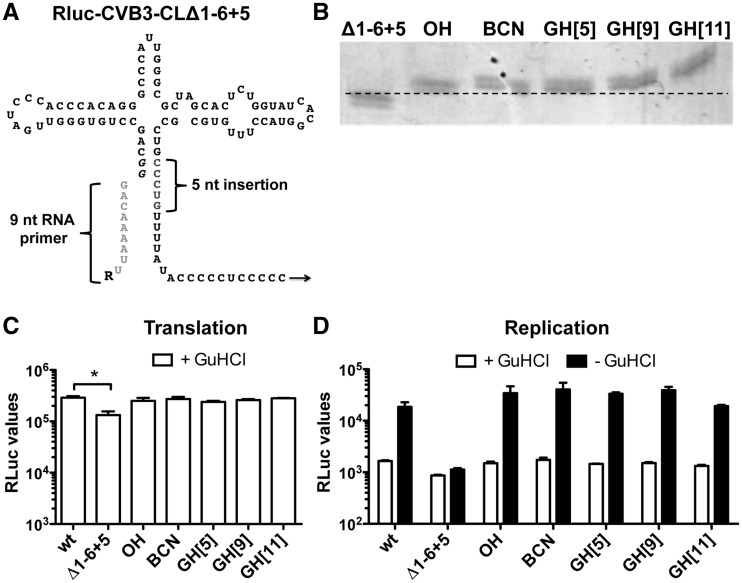
Figure 2.
Base pairing-directed RNA primer ligation to RLuc-CVB3-CLΔ1−6 + 5 genomic RNA. (A) Schematic representation of the CLΔ1−6 + 5-modified CL structure. The 5-nt insertion in the 3′ strand of stem A is indicated in gray. Note that T7 RNA polymerase-transcribed RNA contains two additional guanine nucleotides (italic), which form base pairs with cytosine nucleotides of the 3′ strand of stem A. The 9-nt RNA primer used for base pairing-directed RNA ligation is depicted in light gray, and ‘R’ represents the different 5′ modifications. (B) RNA primer ligation efficiency to genomic RNA possessing the CLΔ1−6 + 5 structure was determined by urea–PAGE analysis of a 250-nt RNase H-digested 5′-terminal fragment. Note that ligation of the RNA primer reduces migration speed of the 250-nt RNase H-digested RNA fragment. (C) Translation of incoming genomic RLuc-CVB3 RNA (wt), RNA holding the mutated CL (Δ1−6 + 5) and RNA ligation products possessing different 5′ modifications (OH, BCN, GH[5], GH[9], GH[11]) were determined by transfection of RNA in HeLa cells in the presence of GuHCl. Eight hours post-transfection, HeLa cells were lysed and RLuc values were determined. Data from a representative experiment are presented as the mean of duplicate ±SD and analyzed using unpaired t-test (* indicates significant difference P < 0.05). (D) RNA replication of the transfected RNA was determined by comparing the RLuc values in the presence of GuHCl inhibitor (+ GuHCl) and in the absence of the inhibitor (− GuHCl). Data from a representative experiment are presented as the mean of duplicate ±SD. Note that the modification of the CL structure (Δ1−6 + 5) hampered RNA replication. However, reconstituting the stem by RNA primer ligation restored RNA replication again.