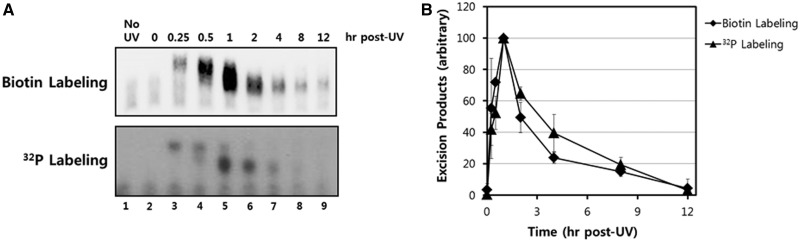
Figure 5.
Sensitivity comparison of the biotinylation and radiolabeling methods. (A) Excised oligonucleotides containing UV damage were recovered from A375 cells at various time points after exposure to 10 J/m2 of UV-C. The purified oligonucleotides were then labeled with biotin-11-dUTP or cordycepin 5′-triphosphate at the 3′-end using terminal deoxynucleotidyl transferase (TdT). Biotin-labeled DNAs were separated on a 12% TBE-urea gel, transferred to a nylon membrane, and detected with HRP-conjugated streptavidin-HRP using a chemiluminescence reagent. Radiolabeled DNAs were separated on a 12% TBE-urea gel and detected by exposing the gel to a phosphorimager screen. (B) Quantitative analysis of the excised repair products. The results from three independent experiments were quantified and are plotted (means ± standard deviation) relative to the maximum.