Figure 1.
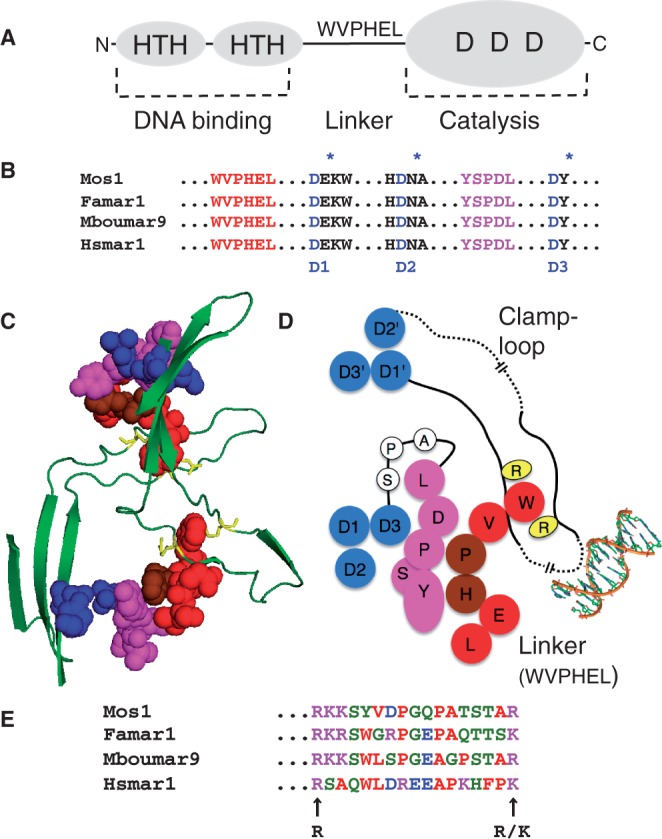
Structural features of the mariner transposase. (A) Transposase has an N-terminal DNA binding domain (amino acids 1–115 approximately) with two helix-turn-helix motifs (HTH). The catalytic domain (amino acids 125–343) has a triad of conserved aspartate residues (DDD), which coordinate the catalytic Mg2+ ions. The domains are connected by a proteolitically sensitive linker region, which harbors the conserved WVPHEL sequence motif. (B) Hsmar1 transposase is aligned with the sequences of three naturally active mariner elements (16–19). In addition to the catalytic triad, mariner has two highly conserved sequence motifs: WVPHEL and YSPDL (20,21). (C and D) The structural relationships between the conserved sequence motifs as they appear in the crystal structure of the Mos1 post-cleavage intermediate (15). A long unstructured ‘clamp-loop' is found inserted between two strands of the core β sheet. The loop extends across the dimer interface and contacts the WVPHEL motif of the opposite subunit. The tryptophan (W) residue is sandwiched between two arginine residues (R). WVPHEL also contacts the YSPDL sequence motif, which is connected to the third active site D residue by a stretch of three residues (A-P-S). Dashed lines indicate breaks in the drawing scale, which is approximate. Also note that the residues in the two dimensional cartoon are arranged to give the most accurate overall picture of the relationships between the structural elements and that some distortions are inevitable. (E) The region of Hsmar1 between the conserved basic residues in the clamp-loop is aligned with the sequences of the three naturally active mariner elements from part B.
