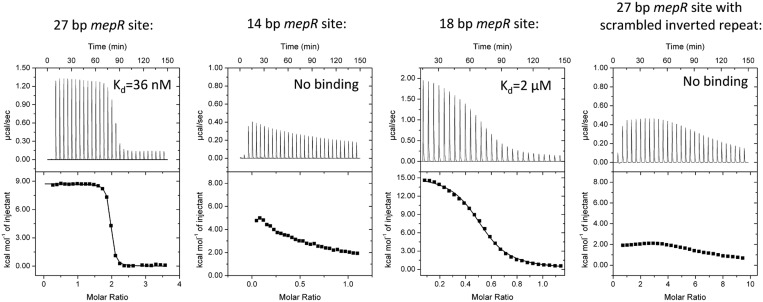
Figure 1.
MepR binding to the mepR operator. ITC thermograms and binding isotherms for four MepR–DNA complexes. In the first and fourth binding experiment, protein (400 and 660 μM, respectively) was injected into DNA (24 and 15 μM, respectively); a stoichiometry of 2 corresponds to 1 dimer of MepR binding to dsDNA. In the second and third experiments, DNA (250 and 400 μM, respectively) was injected into protein (49 and 75 μM, respectively); a stoichiometry of 0.5 corresponds to a single dsDNA molecule binding to a dimer of MepR. The sequences of the oligodeoxynucleotides and fitted thermodynamic parameters are provided in Supplementary Table S1. The data for the 14 bp sequence and 27 bp DNA site with a scrambled recognition helix-binding region demonstrated no binding and were not fit. The molar concentrations of MepR in all experiments were presented per monomer.