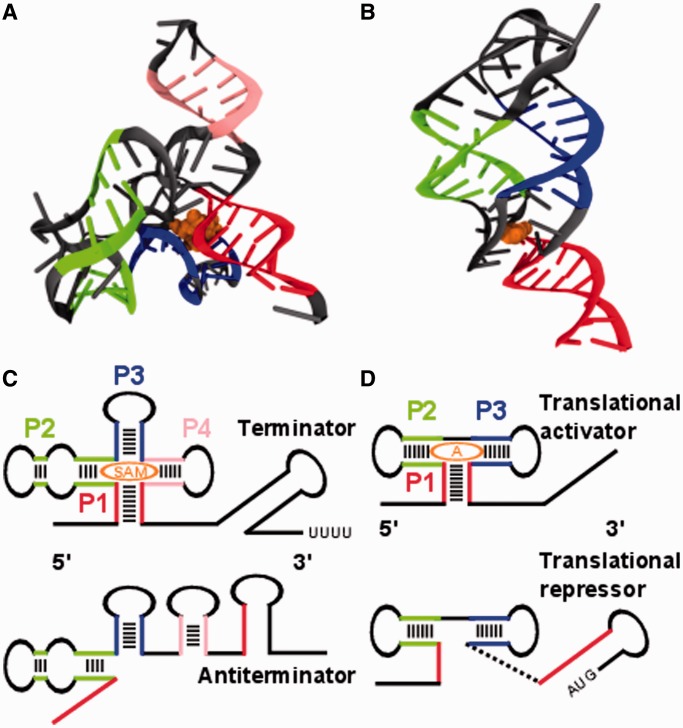
Figure 1.
Tertiary and secondary structures of the SAM-I and adenine riboswitches in ligand bound state. (A) Aptamer region of the SAM-I riboswitch (PDB ID 2GIS): the colored strands indicate elements of secondary structure, helix P1 in red, P2 in green, P3 in blue and P4 in pink. The ligand is shown in orange. (B) Aptamer region of the add adenine riboswitch (PDB ID 1Y26). The same colors are used as in (A) for helices P1 to P3 and ligand. (C) The SAM-I riboswitch consists of two pairs of coaxially stacked helices P1 to P4 connected by a four way helical junction in its ligand bound state. Helix P1 forms in the presence of the ligand and acts as an antiantiterminator allowing the terminator (long-stem loop with downstream sequence of uridines) to fold. In this case, transcription is terminated. (D) The add adenine riboswitch exhibits three helices P1 to P3 in its ligand bound state two of which are coaxially stacked. Helix P1 forms in the presence of the ligand and prevents a translational repressor (initiation codon paired in long-stem loop) from forming.