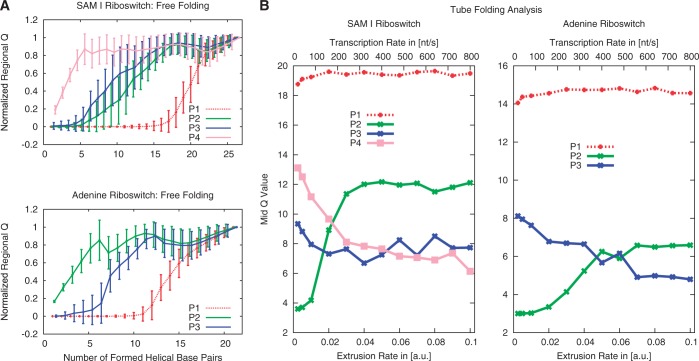
Figure 3.
Folding analysis based on SBM simulations. (A) Folding pathways of the SAM-I and adenine riboswitch for free folding. The plots for each riboswitch are based on 180 folding trajectories, starting from a stretched RNA strand. The mean values and standard deviations are plotted. We can derive the folding order by condensing a substructure’s curve in a single value representing the mid Q-value. (B) Folding events of substructural elements over the extrusion rate. A folding event is characterized by the number of formed helical base pairs at a normalized regional Q-value of 0.5, the mid Q-value. The regional Q-value data are gathered from 80 trajectories for each extrusion rate. The nonlocal helix P1 ties up both ends of the sequence and, therefore, folds last in both riboswitches. Both riboswitches fold in order of appearance in the limit of slow extrusion rates. We investigate a wide range of extrusion rates to cover the natural range of transcription rates. An extrusion rate of 0.0025 corresponds to an estimated transcription rate of ≈ 20 nt/s.