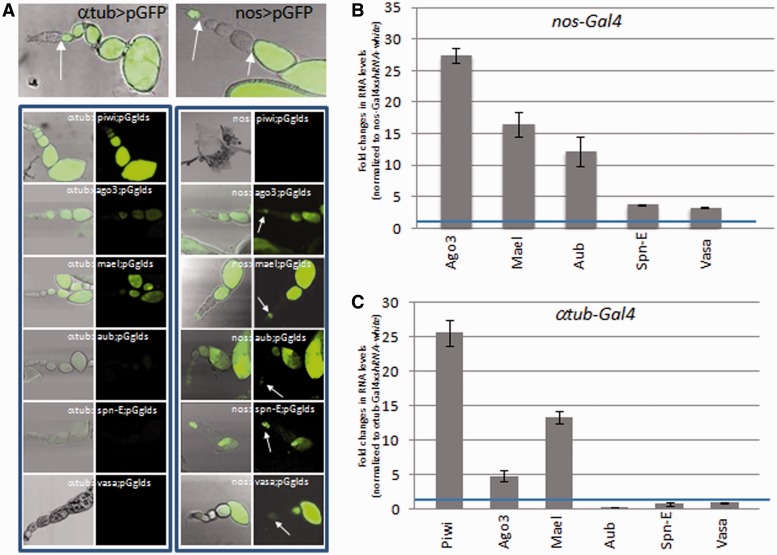
Figure 2.
piRNA pathway components show different temporal requirements for Idefix silencing during Drosophila oogenesis. (A) GFP fluorescence is shown as a readout for pGgIds silencing release using two drivers, αtub-Gal4 (left column) and nos-Gal4 (right column) and RNAi constructs targeting piwi, ago3, mae1, aub, spnE and vasa transcripts. The expression profile of both drivers is presented at the top of each column using a GFP reporter construct (pGFP) and arrows delimit stages where these drivers are active. Ovarioles are oriented with the germarium to the top or left. nos > piwi RNAi results in atrophic ovaries. (B and C) gfp RNA was quantified by qRT–PCR using gfp-specific primers upon RNAi expression targeting the indicated genes and normalized to RNAi against white (the blue line represents no enrichment, value = 1). The transgene pGgIds was driven with nos-Gal4 (B) or α tub-Gal4 (C) (n = 4 biological replicates. Error bars represent SEM). Two transgenic lines pGgIds (s1 and s6) were tested in this experiment.