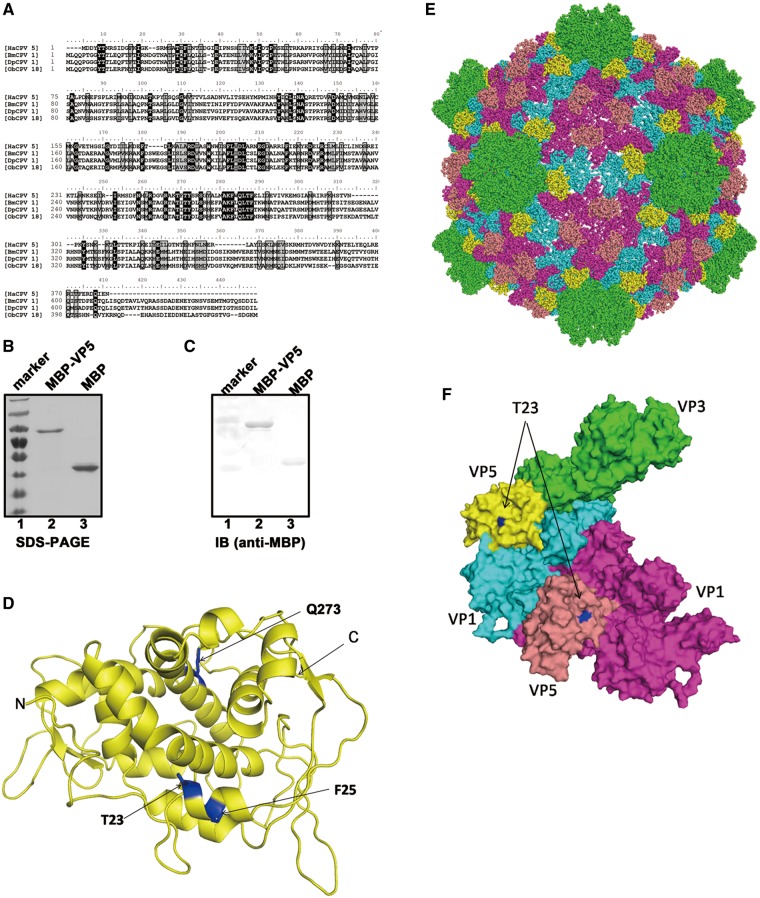
Figure 1.
(A) The amino acid sequence alignment of VP5 proteins of HaCPV-5, BmCPV-1, Dendrolimus punctatus CPV-1 (DpCPV-1) and Operophtera brumata CPV-18 (ObCPV-18). Multiple sequence alignments were generated using ClustalX. (B) Electrophoresis analysis of purified MBP-VP5 and MBP alone. Proteins were loaded onto a 12% SDS-PAGE gel and then visualized via Coomassie blue staining. Lane 1, protein molecular mass markers; lane 2, purified MBP-VP5; lane 3, purified MBP. (C) The purified MBP-VP5 and MBP alone were subjected to SDS-PAGE, followed by western blot analysis with anti-MBP polyclonal antibodies. (D) The 3D structure of HaCPV-5 VP5 was modeled by the HMMSTR/Rosetta server, and drawn by PyMOL as described in ‘Materials and Methods’ section. The three mutation sites as well as N- and C-termini were labeled as indicated. (E and F) The surface representations of modeled VP5, which indicate the orientation of VP5 on the CPV particle (E), or in the asymmetric unit formed by VP1, VP3 and VP5 (F). The T23 on the VP5 is shown in blue. Color-coded by protein subunits, VP3 is in yellow, VP1 has two conformers in cyan and magenta, while VP5 in yellow and salmon. The maps of CPV particle and the asymmetric unit are drawn by PyMOL based on the atomic cryoEM structure of BmCPV capsid (PDB number 3IZX) by replacing BmCPV VP5 with HaCPV-5 VP5.