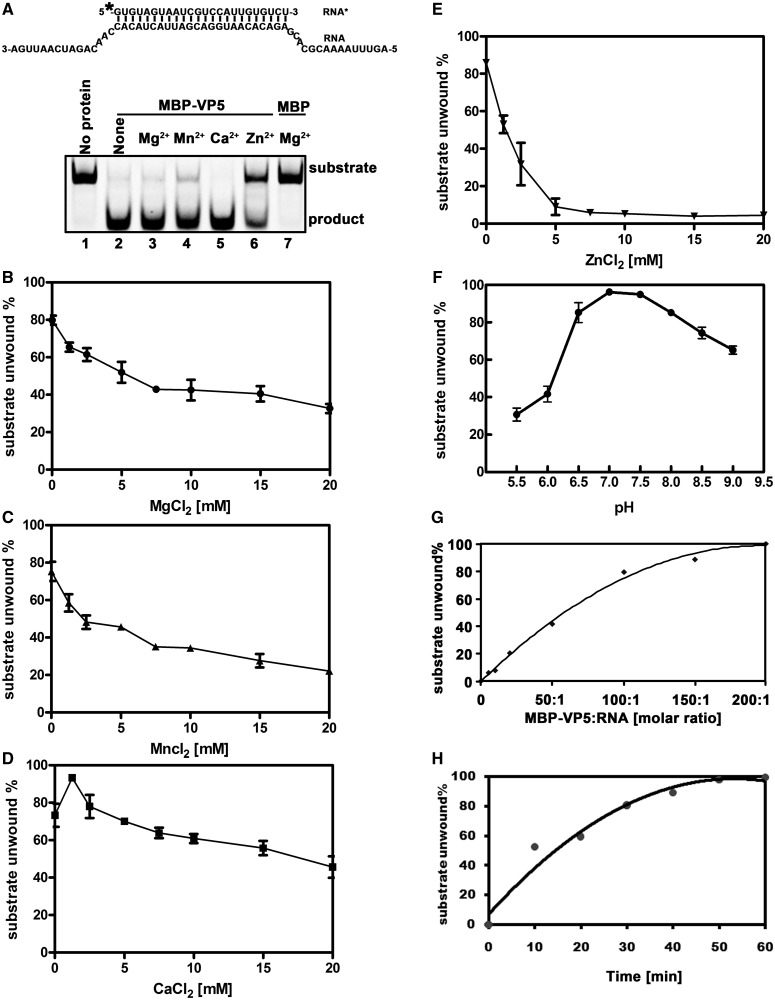
Figure 11.
Optimal biochemical reaction conditions for the RNA helix-destabilizing activity of VP5. (A) Standard RNA helix substrate (RNA1/RNA3; 0.1 pmol) as illustrated in upper panel was incubated with 10 pmol MBP-VP5 in the absence (lane 2) or presence of 2.5 mM indicated divalent metal ions (lanes 3–6) for 60 min. Reaction mixture without protein addition (lane 1) or with MBP alone (lane 7) was used as a negative control. Asterisk indicates the HEX-labeled strand. (B–E) Standard RNA helix substrate (RNA1/RNA3; 0.1 pmol) was incubated with 10 pmol MBP-VP5 in the presence of increasing concentrations of (B) MgCl2, (C) MnCl2, (D) CaCl2 or (E) ZnCl2 as indicated for 30 min. (F) Standard RNA helix substrate (0.1 pmol) was incubated with 10 pmol MBP-VP5 at indicated pH values in the absence of divalent ions for 30 min. (G) Standard RNA helix substrate (0.1 pmol) was incubated with MBP-VP5 at indicated VP5/RNA molar ratios for 30 min. (H) 10 pmol MBP-VP5 was incubated with 0.1 pmol standard RNA helix substrate (at the molar ratio of 100:1) for indicated time intervals. For (B–H), the unwinding activity was plotted as the percentage of the released RNA from the total RNA helix substrate as Y-axis at indicated ion concentrations (B–E), pH values (F), VP5/RNA molar ratios (G) or incubation time intervals (H) as X-axis. Error bars represent SD values from three separate experiments.