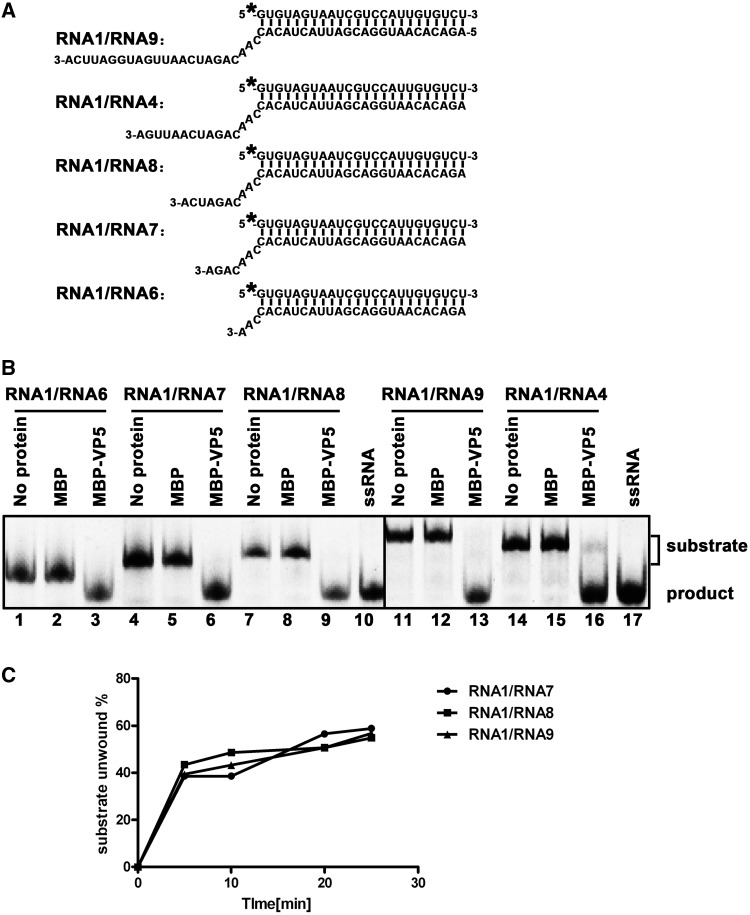
Figure 7.
The length of 3′-tail of RNA helices has no obvious impact on the helix-destabilizing activity of VP5. (A) Schematic illustration of RNA helix substrates with the indicated lengths of 3′-tails. The shorter strand (RNA1) was HEX labeled. (B) Indicated 3′-tailed RNA helix substrate (0.1 pmol) was incubated with 10 pmol MBP-VP5, and the destabilizing activity was determined via gel electrophoresis and scanning on a Typhoon 9200. Reaction mixture without protein supplementation (lanes 1, 4, 7, 11 and 14) or with MBP alone (lanes 2, 5, 8, 12 and 15) was used as a negative control, and ssRNA was loaded to indicate the position of free ssRNA strand in the gel (lanes 10 and 17). (C) Three different RNA helix substrates (RNA1/RNA7, RNA1/RNA8 and RNA1/RNA9) were incubated with 10 pmol MBP-VP5 for 0, 5, 10, 20 and 25 min. The unwinding activity was quantified and plotted as the percentage of the released RNA from the total RNA helix substrate (Y-axis) at each time point (X-axis).