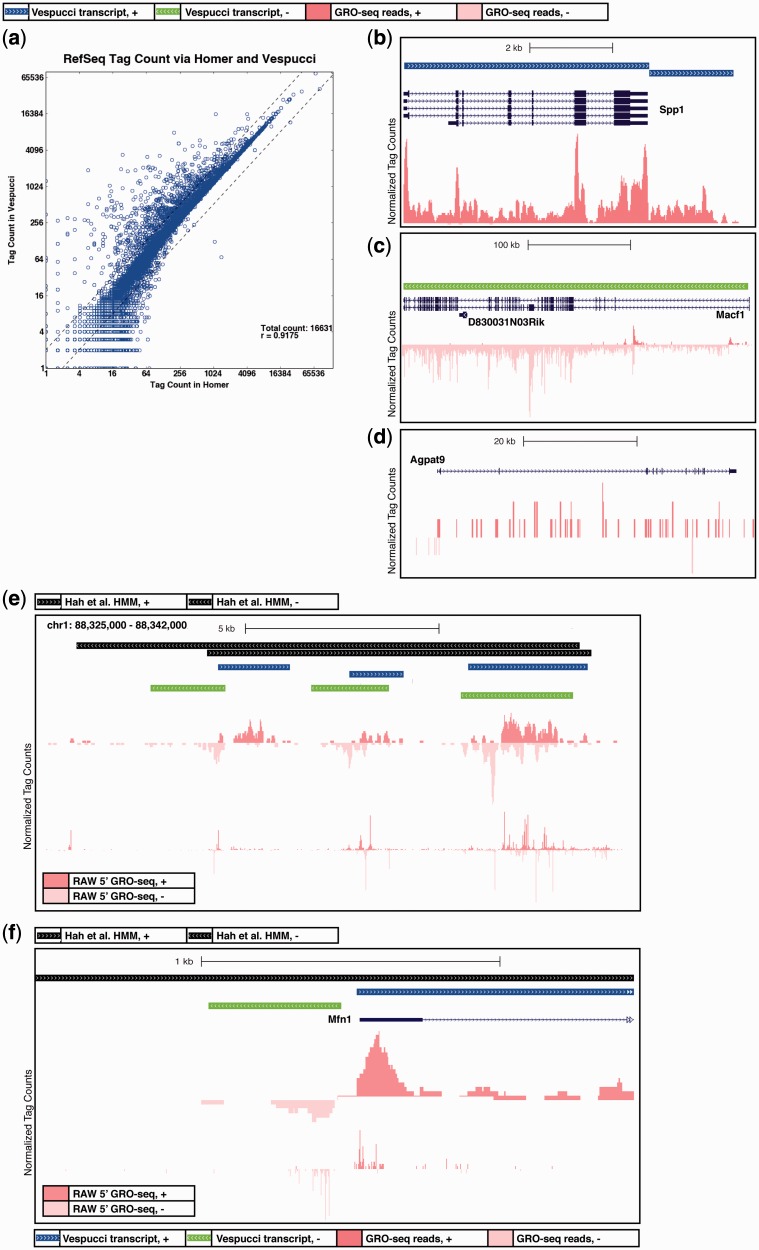
Figure 5.
Vespucci retrieves RefSeq expression levels without losing non-coding RNAs. (a) RefSeq identifiers can be used to compare the tag counts determined by Vespucci at RefSeq genes with the tag counts determined by the HOMER software, which uses a gene-centric approach to sum GRO-seq tags over known genes. The correlation between tag counts is generally good, with deviations from the diagonal attributable to three primary categories of transcripts: (b) Vespucci does not segment transcripts at alternative isoforms, but returns the tags for the whole transcript for each contained isoform. In contrast, HOMER tallies tags within the precise boundaries of each isoform, resulting in discrepancies between the two methods at shorter isoforms, such as the Spp1 gene seen here; (c) as with multiple isoforms, overlapping genes are not segmented by Vespucci, and the tag count for the entire transcript covering Macf1 is associated with the short gene that is overlapping, D830031N03Rik; and (d) genes that have few dispersed tags that cannot be adequately merged yield several smaller transcripts according to Vespucci, whereas HOMER implicitly joins them and counts all that fall along the body of the gene regardless of continuity of transcription. (e) The HMM described by Hah et al. identifies transcripts using a two-state model that calls regions transcribed (black bars) or untranscribed. The HMM identifies many fewer transcripts than Vespucci, in part, because it merges together transcripts called as distinct by Vespucci. Here, three pairs of bi-directional RNAs that are identified as two single units by the HMM. The bottom track shows data from previously published 5′-GRO-seq, a method that detects nascent RNA with a 5′ 7-methylguanylated cap. This method identifies start sites of nascent RNAs genome-wide. The data here, from RAW macrophages, show that Vespucci captures more accurately the separately initiated transcripts. (f) Similarly, some transcripts are called by Vespucci at expression levels too low for the HMM. Here, a paRNA is identified by Vespucci but not the HMM. The bottom track again shows 5′-GRO-seq from RAW macrophages, where the paRNA start site can be clearly seen.