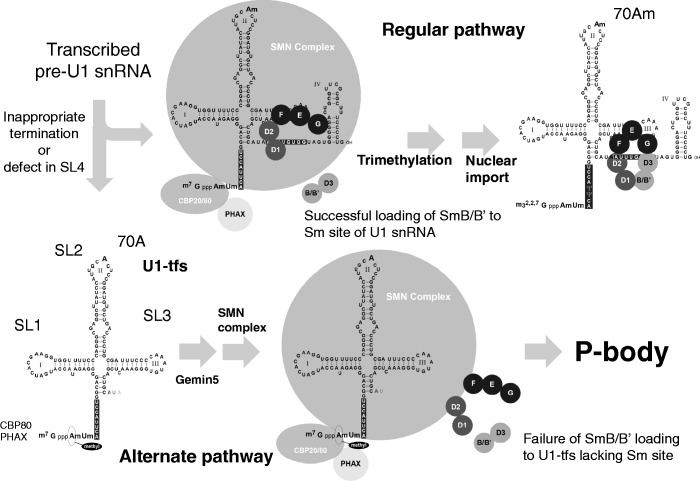
Figure 8.
Proposed alternate pathway for U1 snRNP biogenesis. In an early cytoplasmic stage of the canonical pathway of U1 snRNP biogenesis, the SMN complex containing Gemins 2–8 brings the pentamer of Sm proteins (SmD1, D2, E, F and G) and pre-U1 snRNA together, and then SmB/B′ and D3 are incorporated into the Sm pentamer to form a heptameric Sm ring. Successful formation of the Sm heptamer around the Sm site of pre-U1 snRNA leads to trimethylation of the 5′ cap of pre-U1 snRNA followed by nuclear import to form the mature U1 snRNP with ribose methylation at position 70 (adenosine; 70 Am) of U1 snRNA. In the alternate pathway at an early stage of U1 snRNP biogenesis, some pre-U1 snRNAs with inappropriate 3′ termination or defects in the SL4 region are transformed to U1-tfs with a m7G cap; U1-tfs lack the Sm site and SL4 region and are methylated at position 1 (adenosine) but not at the position-70 ribose. Because U1-tfs lack the Sm site, they are unable to form the Sm heptamer owing to its inability to accept SmB/B′ (and probably SmD3) loading to the Sm pentamer on the SMN complex. Failure of SmB/B′ (and probably SmD3) loading and of formation of the heptameric Sm ring on the pre-snRNA results in an increased number of P-bodies and/or localization of the U1-tfs–SMN complex to P-bodies, which function in RNA surveillance and decay.