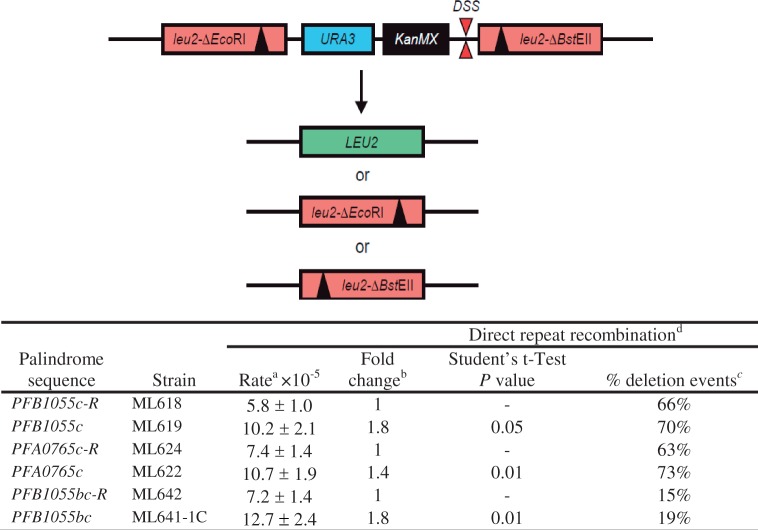
Figure 3.
Effects of predicted DNA secondary structure sequences on mitotic leu2 direct-repeat recombination. The schematic illustrates the assay for spontaneous direct-repeat recombination between two non-functional leu2-ΔEcoRI and leu2-ΔBstEII alleles showing the position of the inserted DSS sequences (DSS). Recombination between the leu2 alleles to produce a functional LEU2 allele leads to prototrophy for leucine (Leu+). The assay scores for Leu+ recombinants generated by single-strand annealing, replication slippage or gene conversion between the leu2 alleles. Single-strand annealing leads to loss of the URA3 gene and uracil autotrophy. The fraction of URA3 deletion events were the same for the pair-wise combinations of strains and their scrambled counterpart, indicating that the 50-mer with the lowest folding free energy stimulated the different types of recombination equally well.
aRecombination rate is presented as events per cell per generation ± standard deviation, as described in ‘Materials and Methods’ section.
bRelative to a randomized sequence (R).
cPercentage of deletion events among Leu+ recombinants.
dStrains harbouring the palindrome sequences PFB1055c (ML619: 5′-TGGTGCCACTGGCAAAAGTGGTGATAAGGGTGCCATTTGTGTGCCACCCA), PFA0765c (ML622: 5′-CAAACACCTGGTGAGAAAACCACCCCACCTAGTGGTACTAACCAGGGTGC) and PFB1055bc (ML641-1C: 5′-GTAAGGACGAAAACGGCAAAAAGCCCGGCTCAAATGCCGACCAAGTCCCC) or their randomized counterparts PFB1055c-R (ML618: 5′-CCTGAAATTGCTGGCTAGGGGTCCTAGATGTGCCCGGGGTAGACCTATAA), PFA0765c-R (ML624: 5′-GCACTGATATGCAAGGAAGCCCCAGCAATCCTCAAAGACGCGAAGCCTCT) and PFB1055bc-R (ML642: 5′-GTATAAGCCTGGAAACCAACAGCGAAAGGCCGAAACCCGCCTACCAAGCG), respectively, were analysed for mitotic direct-repeat recombination at 30°C, as described (49).