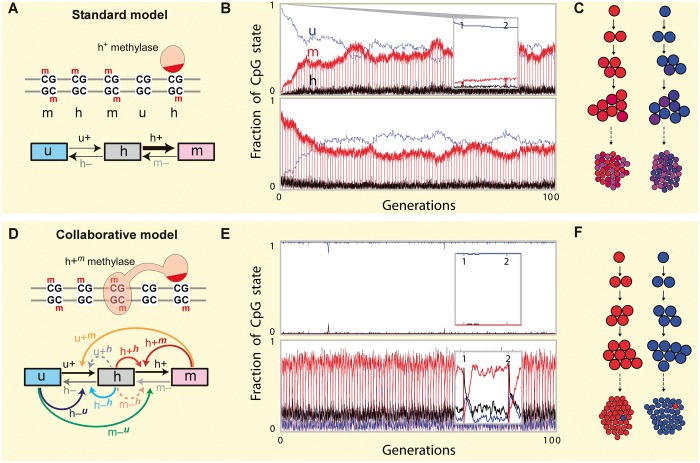
Figure 1.
Standard and collaborative models for DNA methylation. (A) The standard model includes only simple transitions between fully methylated (m), hemimethylated (h) and unmethylated (u) CpG sites. (B) Time course for a typical standard model reaction scheme, tracking the changing fractions of m (red), h (black) and u (blue) CpG sites within the 80 CpG island >100 generations. The island was initialized either in the U state (upper panel) or the M state (lower panel) but converges to an intermediate state u≈m≈1/2 in both cases after ∼20 generations. Replication at the beginning of each generation sets the density of m to 0, which is rapidly restored to nearly pre-replication levels by maintenance methylation, as shown in the inset expanding the period from generations 1–2. (C) In the standard model, CGIs initially in the M state (red) or in the U state (blue) become partially methylated (intermediate shades) in their descendant cell populations. (D) In the collaborative model, methylation and demethylation reactions are affected by the status of nearby CpG sites (curved arrows), e.g. the dark red h+m arrow specifies an h to m transition facilitated by local m sites, which could be achieved by recruitment of a methylase enzyme by m sites. (E) As (B) but for a typical reactions scheme with the collaborative model, showing that a hypomethylated state (U state) or a hypermethylated state (M state) can be stably maintained over many generations. The insets expand the period from generations 1–2, showing the replication-induced disturbance in methylation. (F) The collaborative model can stably maintain bimodal (polarized) methylation states of CGIs in a cell population.