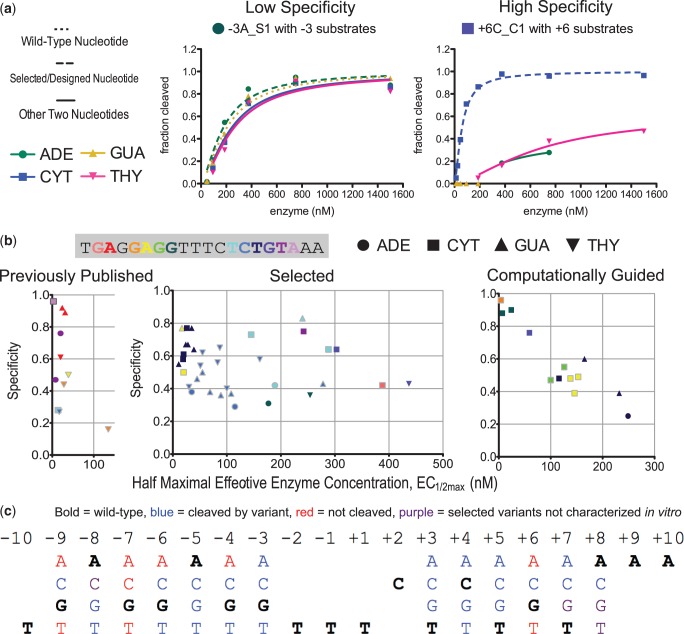
Figure 3.
Combining directed evolution with computational design to successfully select for variants cleaving targets with single base-pair substitutions in the I-AniI target site. (a) Representative data from cleavage assays with each of the four single base-pair substitutions at the targeted position for variants exhibiting high or low specificity. Data for the other I-AniI variants are available in Supplementary Figure S2. (b) Activity and specificity of I-AniI endonuclease variants that cleave single base-pair substitutions (Supplementary Figure S2, Supplementary Table S1). Each point in the graphs is an enzyme variant and is colored to reflect the substituted nucleotide in the cleaved DNA target, both those toward which the enzyme was evolved or the preferred substitution, in the case where an alternate non-WT nucleotide was preferred with over twice the specificity to the target. These three plots include the majority of the 64 variants discussed in this work, with the exceptions of two published enzymes without EC1/2max data available and three selected enzymes with EC1/2max-values of over 500 nM for all base-pairs at the target position. (c) Summary of the single base-pair substitutions in the I-AniI target site that can be cleaved with the engineered variants. Of the 39 substitutions for which selection was attempted—substitutions at −10, +9, +10, and the central four were skipped—variants cleaving 26 substitutions were expressed and characterized (Supplementary Table S1). Blue, sites cleaved by engineered variants; purple, sites for which high-surviving variants were identified but the endonucleases were not expressed; and red, sites for which cleaving variants were not obtained.