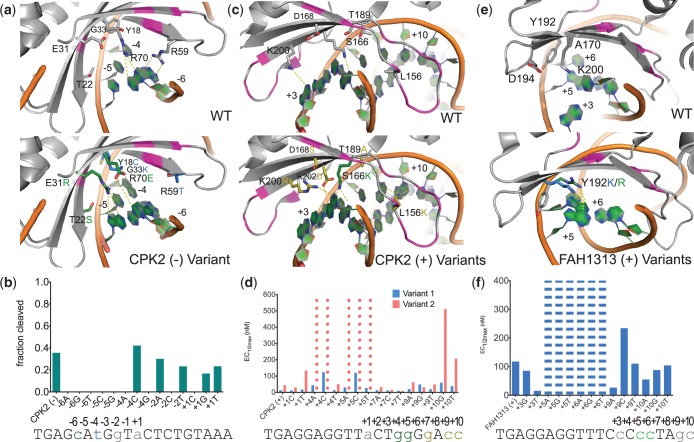
Figure 4.
Combining computation and selection to cleave multiple base-pair substitutions with high specificity. Libraries screened to generate variants, exact mutations tested and the data shown are detailed in Supplementary Table S2 and Supplementary Figure S8. The WT amino acids in the region of interest are shown in gray in the upper portion of panels (a, c and e), and residues predicted to be important for activity and specificity (data in panels (b, d and f)), are shown in the lower portion. Protein backbone positions shown as magenta are supporting positions that were varied in the selection process, but did not yield the dominant contacts. Any substrates not shown in panels (b, d and f) are missing because the process used to generate the target sites utilized a batch method for large numbers of substrates mixed together and these missing constructs were not successfully derived from the group. Bars with dashes indicate that no cleavage was visible at the highest tested enzyme concentration. (a) The (–) half of the CPK2 site contains multiple base-pair substitutions, including −6C and −4T. An enzyme was engineered to target a site with both changes by combining amino acid mutations from variants previously engineered for each single base-pair substitution. Mutations targeting −6C (from −6C_C1) are shown in green and mutations for −4T (from −4C_C1) are shown in blue. (b) Cleavage of the CPK (–) half-site with the CPK2_N enzyme (exact mutations in Supplementary Table S2) and targets containing the indicated base substitution, measured by in vitro cleavage with ∼1700 nM of the CPK2_N enzyme. (c) The (+) half of the CPK2 site contains the adjacent substitutions +4G and +5G, as well as the substitutions +7G, +9C and +10C. The +4G/+5G pocket was targeted before the entire half-site by selecting enzymes from motif-biased libraries, and a library containing the S166K (green) motif produced successful variants that provided a starting place for further selections against the entire (+) half-site. Positions in yellow are contacts that were identified from selections against the CPK2 (+) half-site. For example, the mutation L156K is predicted to hydrogen bond with the two guanines on the reverse strand of the +9C/+10C double base-pair pocket and the D168S mutation caused a significant increase in survival over a D168A-containing variant (Supplementary Table S2). (d) Activity data for two enzyme variants, CPK2_C1 (variant 2) and CPK2_C3 (variant 1), selected from libraries based on the positions shown in panel (c) tested in vitro against the CPK2 (+) half-site and targets with the indicated single base-pair substitution. The two variants differ from each other by two mutations, D168S and T204A, in CPK2_C3 (variant 1) that increase activity over CPK2_C1 (variant 2). (e) The FAH1313 (+) half-site contains the adjacent substitutions +5C and +6C. A library incorporating a motif residue at position 192 was targeted to this double base-pair pocket. Both a lysine and an arginine residue were identified by the computational methods, and surviving variants included both mutations. (f) Activity data for the enzyme FAH1313_Ccc (Table S2), selected to cleave +5C/+6C and incorporating the Y192K motif residue (panel (e)). Data were collected for the full FAH1313 site and all single base-pair substitutions.