Figure 6.
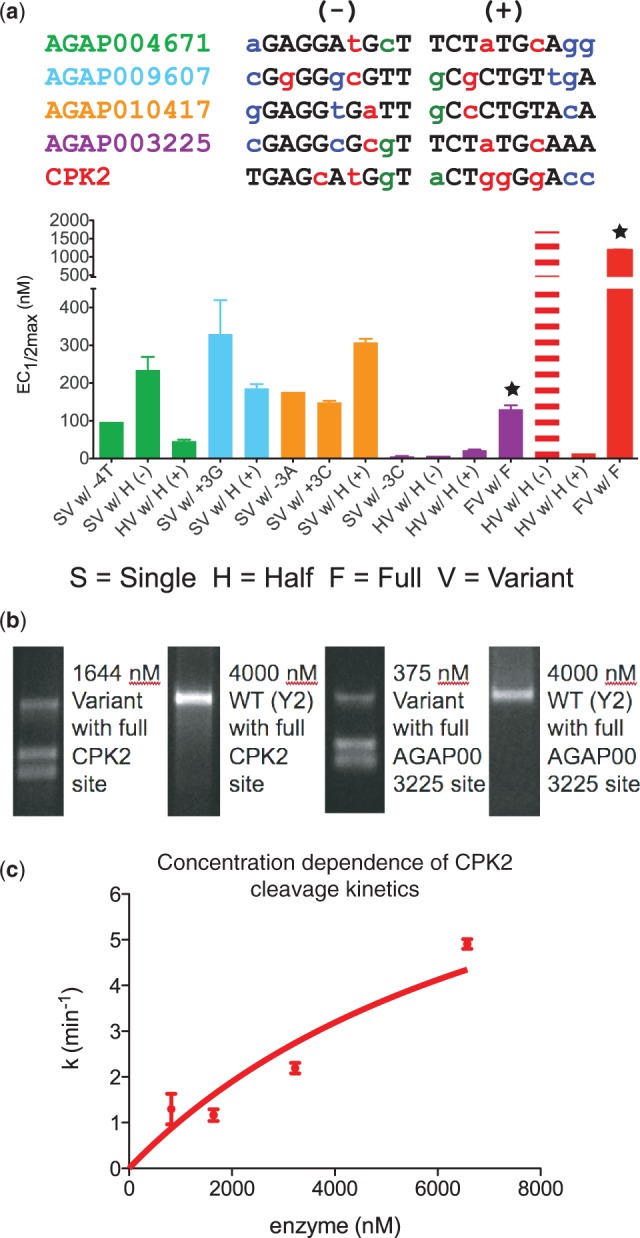
Cleaving full- and half-sites in genes associated with Anopheles sterility and genetic diseases. (a) Activity of variants selected against single base-pair substitutions (SV, single-site variant), half (– or +) (HV, half-site variant) and full-sites (FV, full-site variant) tested against the indicated target with in vitro cleavage assays. The color scheme for base-pair substitutions is the same as Table 1. Single variants are from the set of previously tested variants introduced in Figure 3, and sequences of all tested variants are in Supplementary Table S2. The dashed bars for the CPK2 HV w/ the (–) half-site indicate that an accurate EC1/2max-value could not be collected and this estimate was derived from data in Figure 5. Stars mark the two full variants shown to cleave their respective full-target sites. (b) Gel images showing specific cleavage of full target CPK2 and AGAP003225 sites by engineered enzymes, as well as lack of cleavage by the WT I-AniI scaffold (Y2) at even higher protein concentrations. Specificity for individual positions in the single base-pair changes in the AGAP003225 target and multiple base-pair changes in the CPK2 target are shown in, respectively, Figure 4 and Supplementary Figure S2. (c) Kinetic data for the endonuclease engineered to cleave the full CPK2 target. The rate of target site cleavage by the endonuclease engineered to cleave the full CPK2 target continues to increase above 3 uM indicating a high KM*.
