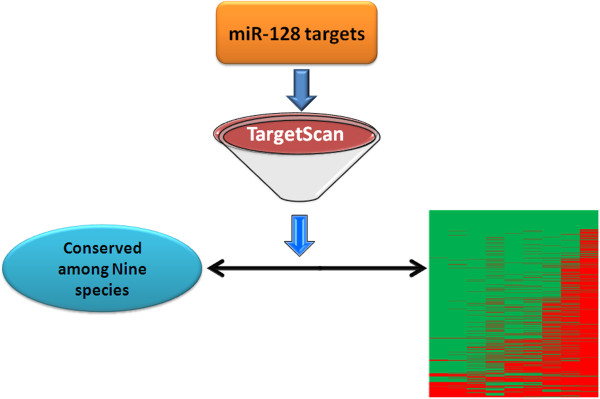
Figure 3.
Strategy for filtering common genes among nine species: Total targets of miRNA-128 have been extracted using TargetScan 5 program for nine species (Human, Mouse, Chimpanzee, Rhesus, Cow, Chicken, Frog, Rat, Opossum). Data was arranged in a tabular format where the union of all genes from the mentioned species were represented as first column in each row (row head). The subsequent columns in first row had species names in them (column head). For every gene, 1 was written under the species where it was found to be present and zero otherwise. This way, a matrix of 1 and zeroes was populated for every gene where 1 means presence and zero means absence. In the last column, sum across the row was taken to count the number of species in which a particular gene was present. We chose only those genes with presence in all nine species. This led to a list of ninety genes which we called high confidence set and were conserved among these species. The total green area specifies ninety common targets whereas red specifies the absence of a particular target in a particular species out of nine species.