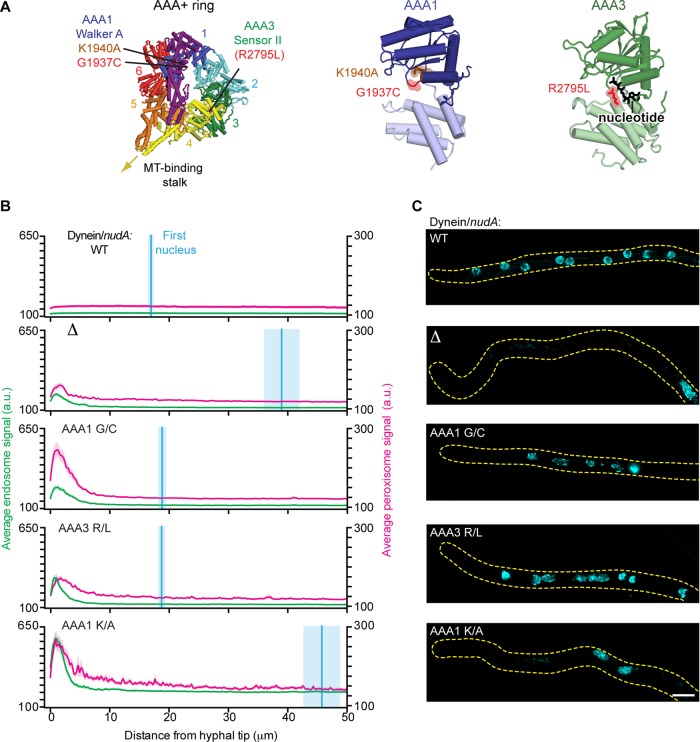
FIGURE 3:
Mutations in dynein that impair the positioning of endosomes and peroxisomes but not nuclei. (A) Structure of the S. cerevisiae dynein motor domain (left; PDB 4AKH; Schmidt et al., 2012). Expected positions of the A. nidulans dynein point mutations (G1937C and R2795L) identified in our screen, as well as the previously characterized K1940A mutation (Zhang et al., 2010) are highlighted. Closeups of AAA1 (middle) and AAA3 (right) are also shown. (B) Plots of endosome, peroxisome, and nuclear distribution in various dynein backgrounds, quantified as described in Figure 2D. DyneinΔ, dyneinAAA1 G/C, dyneinAAA3 R/L, and dyneinAAA1 K/A hyphae all showed defects in endosome and peroxisome distribution, as there was a pronounced peak of fluorescence intensity for endosomes and peroxisomes in the hyphal tip compared with wild-type hyphae. Nuclear distribution was perturbed in dyneinΔ and dyneinAAA1 K/A hyphae but not in dyneinAAA1 G/C, dyneinAAA3 R/L, or wild-type hyphae. The mean ± SEM distances from the distal nucleus to the hyphal tip were 16.95 ± 0.57 μm (n = 52) in wild-type, 38.97 ± 3.12 μm (n = 31) in dyneinΔ, 18.81 ± 0.78 μm (n = 46) in dyneinAAA1 G/C, 18.71 ± 0.71 μm (n = 36) in dyneinAAA3 R/L, and 45.75 ± 3.22 μm in dyneinAAA1 K/A (n = 45) hyphae. The distal nucleus position in dyneinAAA1 G/C and dyneinAAA3 R/L hyphae was not statistically different from that in wild-type hyphae (p > 0.05, unpaired t test), whereas the nuclear distribution in dyneinAAA1 K/A and dyneinΔ hyphae was statistically different from that in wild-type hyphae (p < 0.0001, unpaired t test). (C) Representative micrographs showing nuclear distribution in A. nidulans hyphae in the various dynein backgrounds. Scale bar, 5 μm.