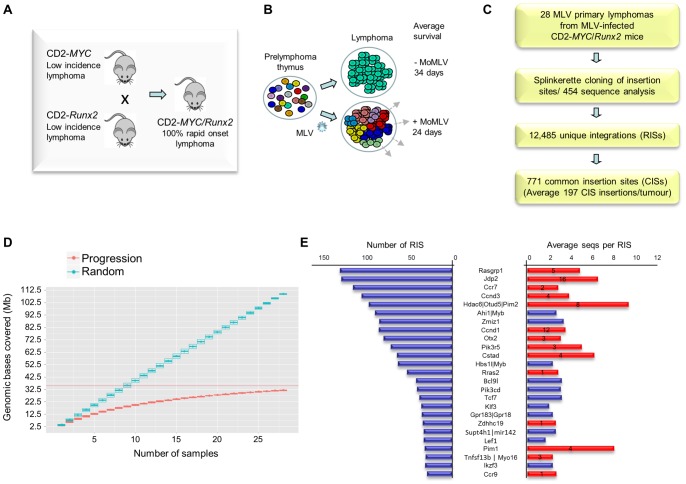
Figure 1. (A) Features of the system and experimental design of the RIM/DS progression screen (see also Figure S1).
Mice carrying CD2-MYC or CD2-Runx2 transgenes each develop a low incidence of lymphoma, while double transgenics develop lymphomas early with 100% penetrance. (B) Infection of double transgenic mice with MoMLV increases the rate of lymphoma development and the clonal complexity of the resulting tumours. (C) Flowchart of sequencing analysis: Splinkerette clones from 28 double transgenic tumours were sequenced by Roche 454 to identify 12,485 unique retroviral insertion sites (RISs). Gaussian kernel convolution statistical analysis identified 771 common insertion sites (CISs). (D) Saturation analysis of common insertion sites from the 28 MoMLV-accelerated thymic lymphomas. The number of genomic bases covered by predicted CISs increases as the number of samples used increases. The increase is linear if RIS are randomly distributed (upper) but approaches saturation in our real dataset (lower), indicating that 28 samples is sufficient to identify almost all positively selected CISs in this experimental system. (E) The 25 most frequently targeted CISs ranked by number of individual RISs. The right-hand panel shows the average number of reads for RIS. Red bars denote those detected in a previous shotgun cloning screen, with numbers denoting the number of clones detected [9]. A positive correlation (R = 0.56) was noted between with the number of reads/RIS and likelihood of detection by the lower-powered shotgun cloning methodology.