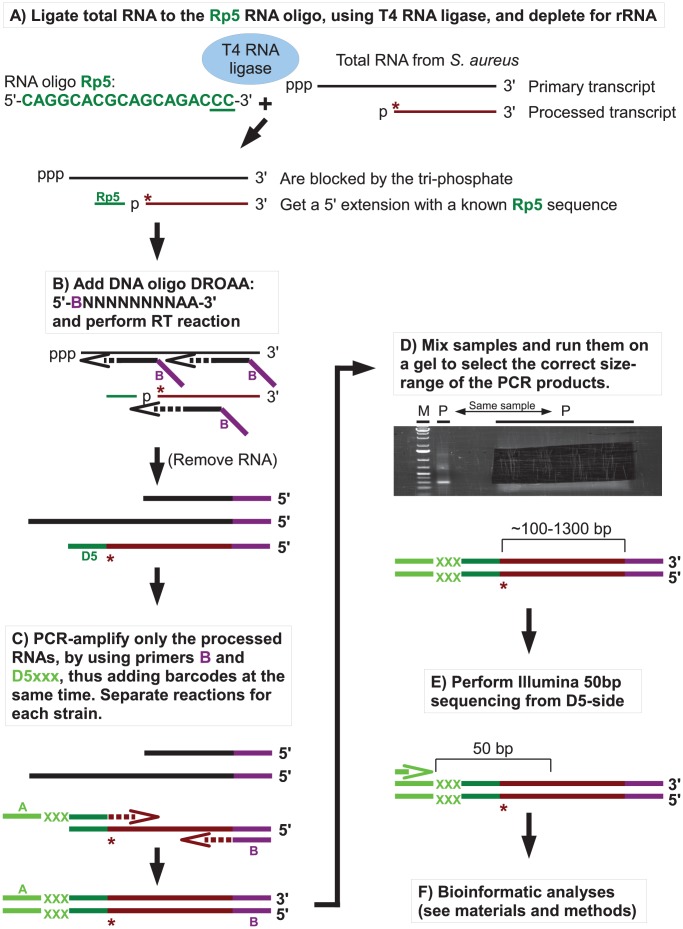
Figure 3. Flowchart showing the mapping of 5′ RNA ends with the EMOTE assay.
The 5′-bases of mono-phosphorylated RNA (asterisks) were identified using the following technique. Thin and thick lines denote RNA and DNA, respectively. Open arrows indicate primer elongation. A) The RNA oligo Rp5 (green) is ligated to total RNA from each strain, adding a known sequence to all mono-phosphorylated RNAs (brown), and excluding tri-phosphorylated RNAs (black). The majority of 16S and 23S rRNA are removed by hybridisation to magnetic beads. The two underlined cytidine nucleotides at the 3′-end of Rp5 will be explained below. B) Reverse transcription is performed with a semi-random primer (DROAA), which cannot anneal to the Rp5 sequence. The 5′-ends of the cDNA will all have the sequence of Illumina adaptor B (purple), but only cDNA made from Rp5-ligated RNA will end in a known sequence (D5), which is complementary to Rp5 (green). C) The cDNA from each strain is amplified using one primer, B, that anneals to the Illumina adaptor B (purple), and a second primer, D5xxx, that anneals to D5 (green) and adds the Illumina adaptor A sequence (light green) as well as a short bar-code (light green “XXX”) which is unique to each strain. The D5xxx primers only have the first 16 nt of Rp5, so any PCR product where the D5xxx primers have annealed to non-Rp5 sequence, will not include base 17 and 18 of Rp5. D) The PCR products from all the strains are mixed together and run on an agarose gel, whereupon the appropriate size-range of the smear (P-lanes) is extracted. E) Illumina sequencing of 50 bases from the D5-side then reveals the bar-code (light green, identifying from which strain the original RNA came), the Rp5 sequence (green), the first base in the original mono-phosphorylated RNA (asterisk), and the subsequent 23 bases (brown), allowing an unambiguous mapping of the 5′-end of each detected RNA. F) Bioinformatic analyses to (I) use the bar-code sequence to assign each read to the correct strain. (II) Verify that the two cytosine bases at the 3′-end of the Rp5 sequence are present to remove reads that originate from a mis-priming of the D5xxx primer. (III) align the 24 bases to the genome, to determine the exact position and orientation of each 5′-base. (IV) Tabulate and analyse the data (see materials and methods).