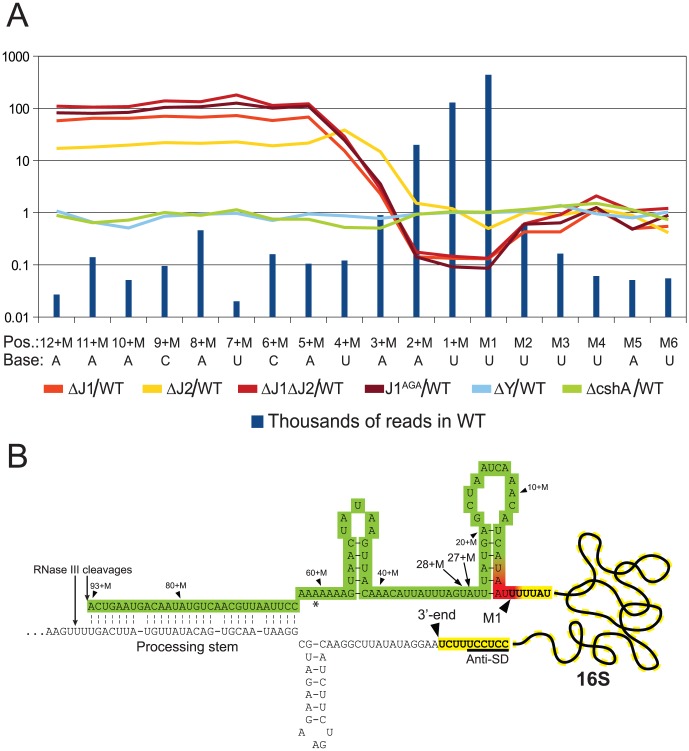
Figure 4. Enrichment of immature 16S rRNA in RNase J mutants.
A) The curves show the number of reads mapping to each position of the immature 16S rRNA (mature 16S starts at position M1) in the mutant strains, divided by the number of reads mapping to the same position in the WT data. Data from RNase Y and cshA deletion mutants [21] have been included as controls. The number of reads for each strain has been normalised to the sum of reads mapping to positions between the 16S rRNA RNase III processing site [25] and the 3′-end of the mature 16S rRNA. Blue bars indicate how many thousands of reads mapped to each position in the WT data-set. B) Overview of the region important for the maturation steps after RNase III has cleaved the processing stem. Positions of 5′-ends that accumulate in the RNase J mutants are indicated in green, and red positions are much less abundant in the RNase J mutants compared to the WT strain. No significant changes are observed at the yellow positions. M1 denotes the first nucleotide of the mature 16S rRNA. The Anti-Shine-Dalgarno (Anti-SD) and the 3′-end of the mature 16S rRNA are indicated. 5′-ends at positions 28+M and 27+M are relatively frequently observed (Table S3), and might be a hotspot for the ribonuclease that cleaves between 93+M and M1, prior to trimming by RNase J (see discussion for details). The asterisk indicates where two of the five 16S rRNA genes in S. aureus have one base less.