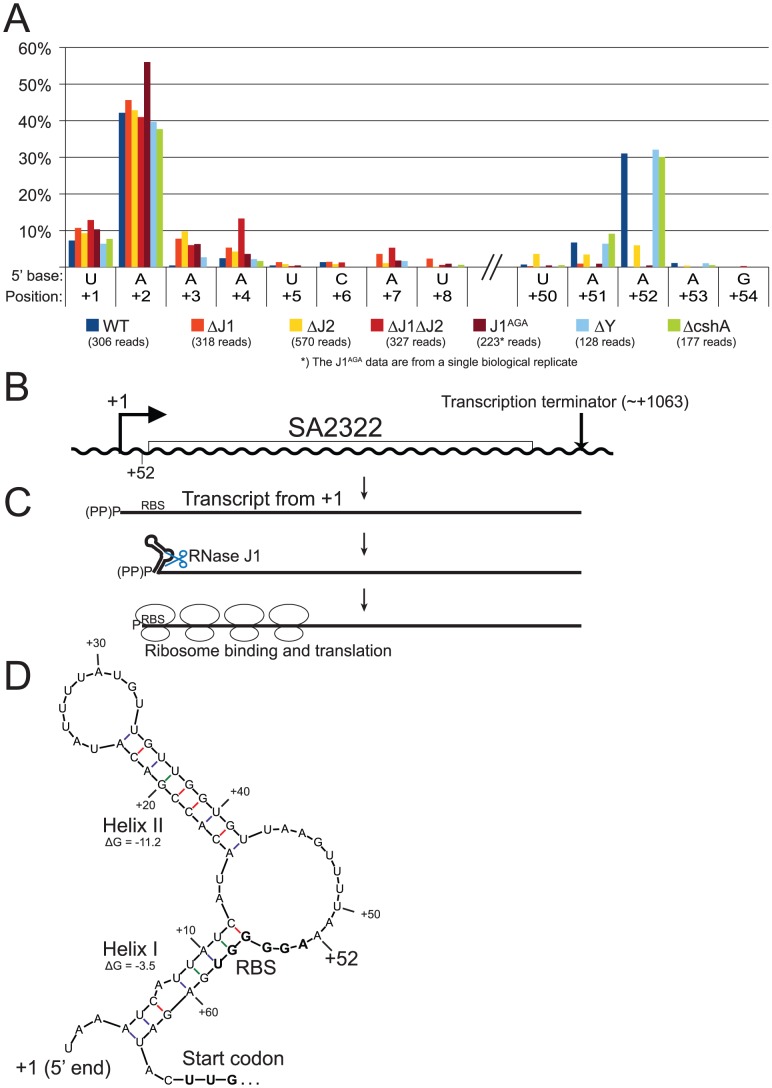
Figure 6. mRNA maturation by RNase J reveals a potential regulation of translation.
A) Histogram showing the percentage of reads mapping to a given position, out of the total number of reads mapping to the SA2322 transcript in each strain (shown in parentheses). Only positions of interest are included, but the full data-set can be found in Table S7. +1 and +2: The putative transcription start sites. +52: A major detected RNA species in the WT, ΔY and ΔcshA, however it is absent from the RNase J1 mutants and strongly reduced in the ΔJ2 strain. B) The SA2322 locus with important positions indicated. C) A schematic view of the fate of SA2322 transcripts. A newly formed transcript can form a secondary structure, shown in panel D, which partially sequesters the ribosome binding site (RBS). RNase J can shorten the transcript by 51 nt, and is presumably blocked from further exonucleolytic digestion by ribosomes binding to the RBS. (PP)P indicates a mix of tri- and mono-phosphorylated RNA, generated by pyrophosphohydrolases. D) Predicted secondary structure at the 5′-end of the SA2322 transcript. ΔG values predicted by the mfold algorithm are in kcal/mol. RBS and start codon are indicated in bold.