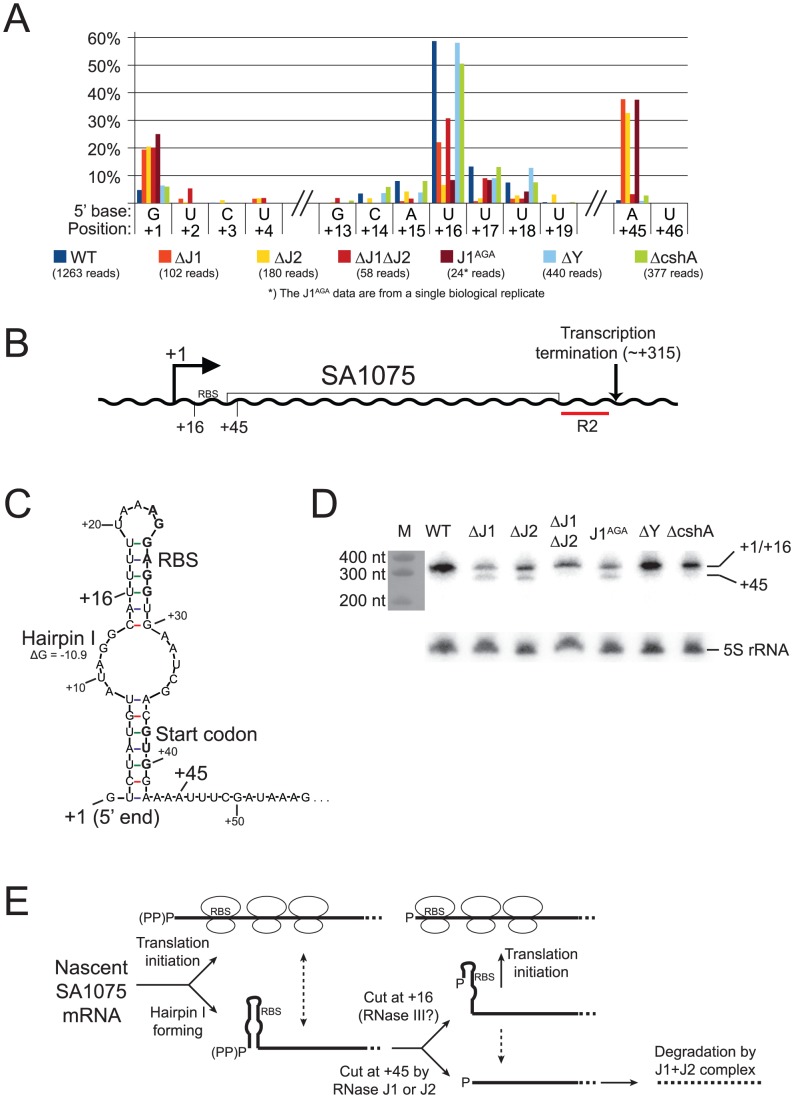
Figure 7. SA1075 mRNA inactivation by RNase J competes with translation initiation.
A) Histogram showing the percentage of reads mapping to a given position, out of the total number of reads mapping to the SA1075 transcript in each strain (shown in parentheses). Only positions of interest are included, but the full data-set can be found in Table S8. +1: The putative transcription start site. +16: The major detected RNA species in the WT, ΔY and ΔcshA. +45: Position where ΔJ1ΔJ2 differs from ΔJ1, ΔJ2 and J1AGA, and appears similar to WT. B) Important positions indicated on the SA1075 gene. +1: Transcription Start Site. RBS: Ribosome Binding Site. R2 indicates the probe used for the Northern blot in panel D. C) Hairpin structure predicted by the mfold algorithm, which sequesters the RBS and start-codon (shown in bold). No secondary structure was predicted for the 50 nucleotides downstream of position +45. D) Northern blot of the SA1075 transcript, using probe R2. The +1 and +16 RNA species are not resolved, and can be seen as a single band, however the +45 species is clearly visible in the ΔJ1, ΔJ2, and J1AGA strains. The marker was stained with methylene blue and photographed. As a loading control, the Northern blot was stripped and re-probed to detect the 5S rRNA. E) The proposed model for determining the fate of SA1075 mRNA via competition between RNase J, ribosomes and the nuclease that cleaves at position +16. (PP)P indicates a mix of tri- and mono-phosphorylated RNA, generated by pyrophosphohydrolases. Nascent SA1075 mRNA can either be occupied by ribosomes, binding to the RBS, or form Hairpin I which sequesters the RBS. Ribosomes will shield position +45 from RNase J, but the hairpin will not. If cleavage at position +16 occurs before RNase J has cleaved at position +45, then the RBS will be liberated from Hairpin I, and ribosomes can initiate translation. If ever the +45 cut is made by RNase J, then the mRNA, which no longer has RBS or start-codon, is immediately degraded (possibly by the RNase J1+J2 complex). Either RNase J1 or RNase J2 can perform a cleavage at position +45. The loss of both RNases prevents the +45 RNA species from being generated, thus explaining why the WT and the ΔJ1ΔJ2 strains appear similar in panel A (see discussion for details and other potential explanations).