Table 3. Comparison of the 3D segmentation quality.
| Algorithm | RI | JI | HM | NSD(x10) | Split | Merged | Added | Missing | t [s] | t [s]* |
| OTSU | 97.35 | 6.00 | 22.82 | 5.72 | 0.00 | 3.27 | 0.87 | 0.00 | 0.49 | 0.44 |
| OTSUWW | 97.57 | 6.03 | 3.80 | 1.12 | 0.13 | 0.00 | 0.00 | 0.00 | 2.57 | 2.48 |
| GFT | 88.06 | 3.57 | 6.81 | 6.25 | 0.10 | 1.57 | 6.53 | 1.87 | 15.51 | – |
| GAC | 95.06 | 6.40 | 7.41 | 2.52 | 0.00 | 1.13 | 0.00 | 0.77 | 5.92 | – |
| GC | 97.78 | 6.37 | 5.66 | 1.69 | 1.34 | 0.07 | 0.00 | 0.00 | 5.92 | – |
| TWANG | 93.82 | 4.94 | 6.62 | 2.41 | 0.00 | 0.00 | 0.00 | 1.37 | 3.72 | 1.08 |
Comparison of the segmentation quality on simulated 3D benchmark images by Svoboda et al. (HL60 cell line, low SNR, 75% clustering probability) [35]. For quality assessment we used the Rand Index (RI), the Jaccard Index (JI), the Hausdorff Metric (HM) and the Normalized Sum of Distances (NSD) as defined in [33]. Besides correct segmentations, nuclei can be split, merged, erroneously added or are missing. The listed values are the arithmetic mean values of 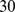 processed 3D benchmark images. Performance of the algorithms was tested without using threads and with 8 threads where possible (indicated by *).
processed 3D benchmark images. Performance of the algorithms was tested without using threads and with 8 threads where possible (indicated by *).
