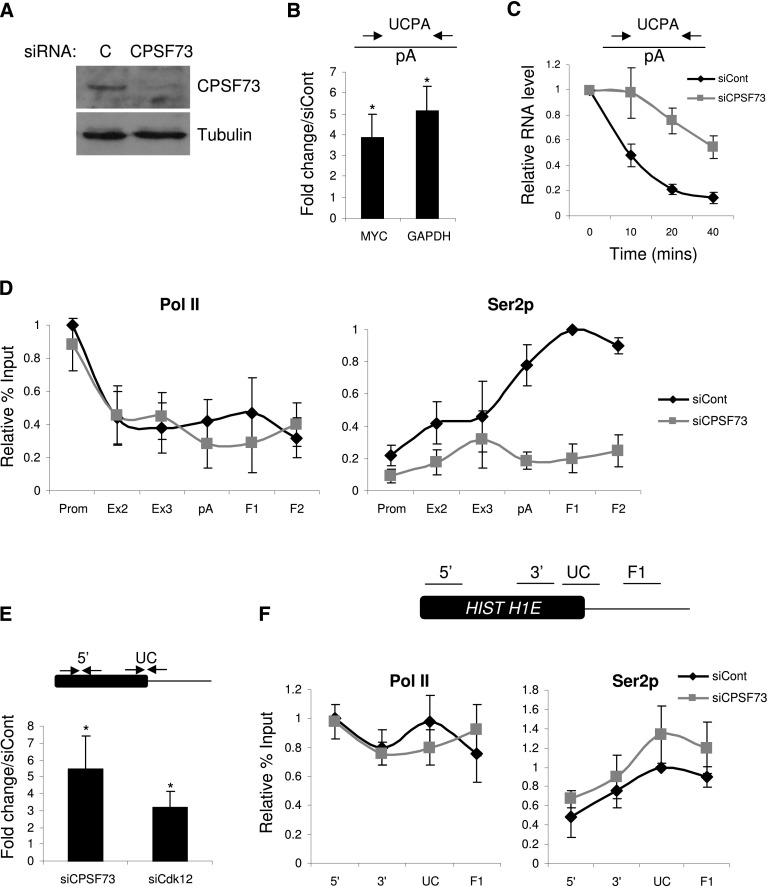
Figure 4.
(A) Western blotting of protein extracts from cells treated with control or CPSF73 siRNAs. The top panel shows CPSF73 protein, and the bottom panel shows Tubulin loading control. (B) Real-time PCR analysis of RNA not yet pA-cleaved (UCPA) from the MYC and GAPDH genes in control and CPSF73-depleted samples. Quantitation shows a fold change in CPSF73-depleted cells relative to control cells normalized to the terminal intron–exon junction. (*) P < 0.05. (C) Act D time-course analysis of non-pA-cleaved (UCPA) RNA in control or CPSF73-depleted cells. RNA levels were normalized to those present at time 0. (D) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the MYC gene in control or CPSF73-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. (E) Real-time PCR analysis of histone H1E RNA 3′ end processing in control, CPSF73-depleted, or Cdk12-depleted samples. The primer pairs used are indicated on the accompanying diagram. Quantitation shows a fold change relative to control cells normalized to signal over the Histone gene body (5′) (*) P < 0.05. (F) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the histone H1E gene in control or CPSF73-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. The diagram shows the positions of amplicons. All error bars represent standard deviation from at least three biological replicates.