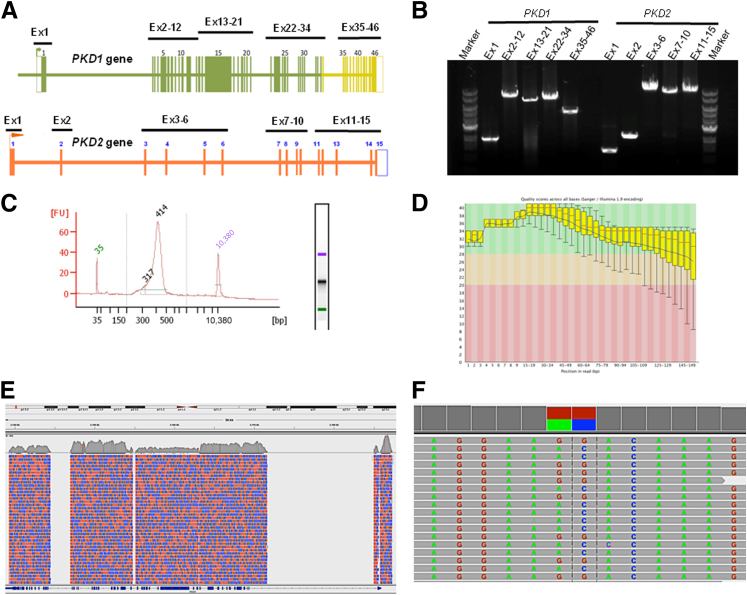
Figure 1.
Visualization of the NGS workflow. PKD1 and PKD2 genes were individually amplified as 10 locus-specific LR-PCR products (1.4 to 10.9 kb in size), with all coding regions and most intronic regions covered, in total, an approximately 68.0 kb genomic region. A: Map of the PKD1 and PKD2 genes showing the position of the 10 pairs of primers used for LR-PCR amplification of the coding regions. The highlighted green and yellow regions correspond to the duplicated and single-copy sequences of PKD1, respectively. B: Amplification quality was verified using agarose gel electrophoresis. C: LR-PCR products from each patient were pooled together at equimolar ratio, followed by fragmentation and library preparation. The finished libraries were quantified and batched together at equimolar amounts in groups of 25 patient samples and were assessed for quality by a high-sensitivity chip using an Agilent Bioanalyzer instrument. D: The pooled libraries were sequenced on an Illumina MiSeq platform. The raw sequencing reads were sorted by bar code first and then were subjected to quality control analysis before proceeding with the mutation analysis. The quality score (Phred-like score) is shown at each position of the reads. E: Reads were then mapped back to the PKD1/PKD2 loci of human genome assembly hg19 using the BWA program. In this example, PKD1 sequencing coverage is shown using the Integrative Genomics Viewer (Broad Institute, Cambridge, MA). Red areas, reads from the plus DNA strands; blue areas, reads from the minus strands. F: Variant callings were made by the GATK software package and were visualized using the Integrative Genomics Viewer. Ex, exon; FU, fluorescence unit.