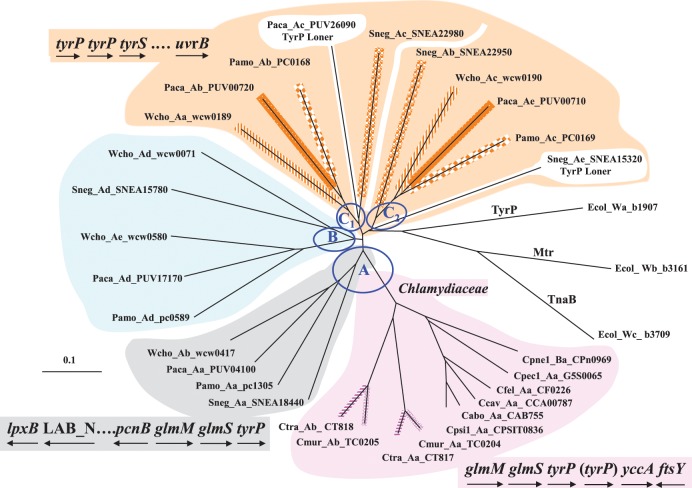
Figure 5.
Multiplicity of the TyrP transporter for Trp in the Chlamydiales Order. The three E. coli paralogs for Trp transport are shown at the right. Three main ortholog clusters of TyrP proteins (designated as A–C) are evident, together with an admixture of a few additional paralogs. Cluster A contains one TyrP protein from Wcho, Paca, Pamo, and Sneg – as well as the divergent set of proteins from the Chlamydiaceae. Ctra and Cmur each have two paralogs, which appear to have occurred independently via gene duplications that followed the speciation divergence. At the lower left is shown a suggested ancestral gene neighborhood which can be compared to the Chlamydiaceae gene neighborhood shown at the lower right. In the latter gene neighborhood inclusion of “(tyrP)” is relevant only to the gene duplicate present in the Cmur/Ctra pair. The Chlamydiaceae have no representation in ortholog clusters B and C. Cluster B possesses members from Wcho, Sneg, Paca, and Pamo – with Wcho having an additional paralog member. Cluster C is comprised of tandem tyrP genes (see the gene neighborhood at the upper left) which encode the four sets of ortholog pairs shown (with matched line patterns). Thus, duplication of the ancestral Group-C ortholog in the common ancestor of Wcho, Sneg, and Paca/Pamo generated the paralog sets that are positioned within Group C1 and C2. In addition Paca and Sneg possess a third paralog belonging to cluster C; the genes encoding these are unlinked to the aforementioned tandem genes (hence their gene products being designated as “TyrP Loners”). Abbreviations for organisms populating the Chlamydiaceae: Ctra, Chlamydia trachomatis; Cmur, C. muridarum; Cpsi, C. psittaci; Cabo, C. abortus; Ccav, C. caviae; Cfel, C. felis; Cpec, C. pecorum; and Cpne, C. pneumoniae. Other Chlamydiales organisms are abbreviated as given in Figure 4.