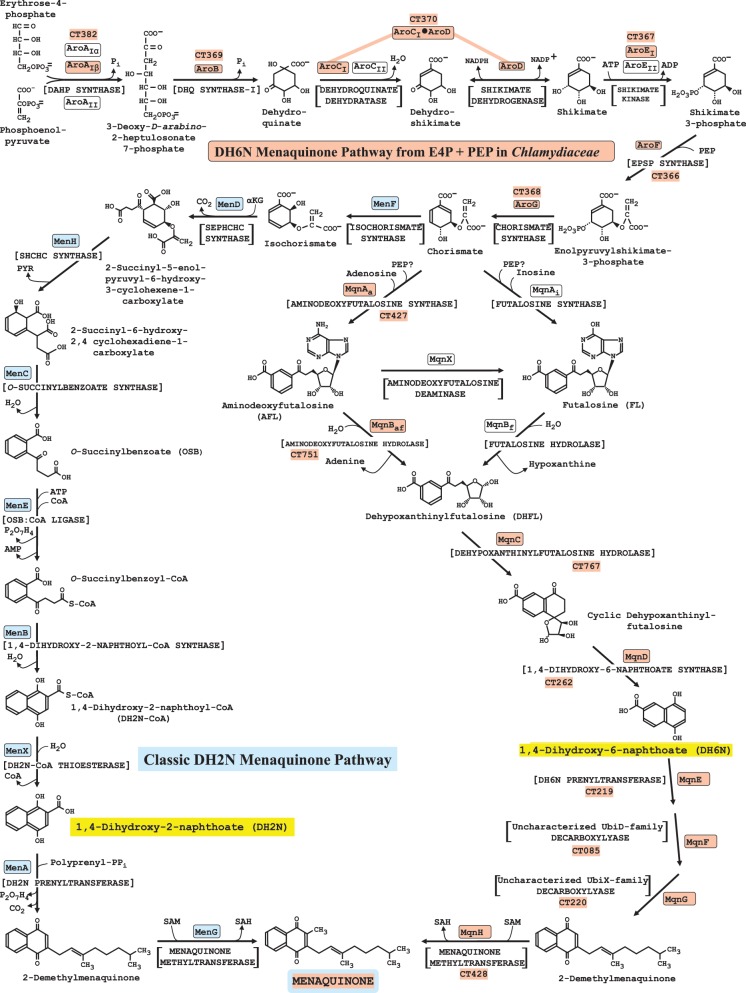
Figure 6.
Variant menaquinone pathways in nature. The composite diagram shows the biochemical variations to menaquinone that are so far known to exist in nature. The proposed 15-step Chlamydiaceae pathway, which includes the 7-step pathway to chorismate, is indicated with orange boxes surrounding the enzyme acronyms. For reference, the encoding CT gene numbers for C. trachomatis D/UW-3/CX are also shown. Multiple acronyms are indicated for the three enzyme steps in the chorismate pathway that can be performed by distinct sub-homolog types or by non-homologous isofunctional analogs (see http://www.aropath.lanl.gov/ for the logical system of acronym assignment used). AroCI and AroD are domain components of a single protein encoded by fused genes. The classic dihydroxy-2-naphthoate (DH2N) pathway from chorismate, generally known as the isochorismate pathway of menaquinone biosynthesis, is shown with acronyms in blue boxes at the left. The alternative DH6N pathway shown at the right was originally called the futalosine pathway (Dairi, 2009, 2012), but futalosine has proven to not necessarily be used as an intermediate because of alternative early-pathway steps that exist. Therefore, we refer to this as the DH6N pathway. The nine enzymes of the DH6N pathway in S. coelicolor are encoded by SCO4506, SCO4662, SCO4327, SCO4550, SCO4326, SCO4491, SCO4490, SCO4492, and SCO4556. The DH2N and DH6N structures, for which the pathways are named, are highlighted in yellow.