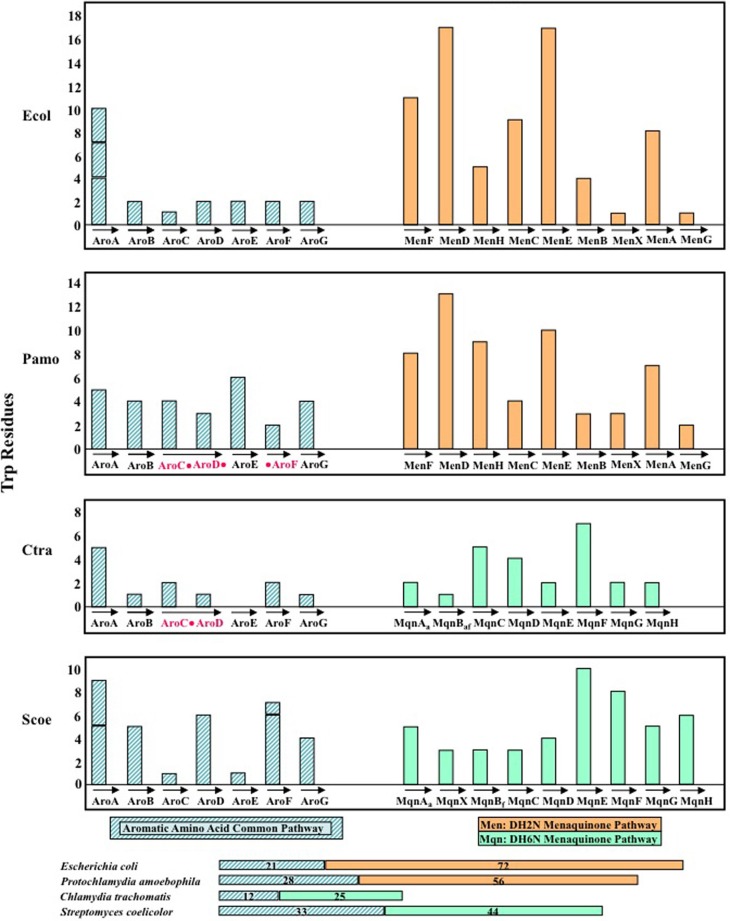
Figure 7.
Comparison of the Trp content of the chorismate/menaquinone pathway in Chlamydia trachomatis (Ctra), its close relative Protochlamydia amoebophila (Pamo), the classic bacterium Escherichia coli (Ecol), and Streptomyces coelicolor (Scoe). Pamo and Ecol both utilize the classic DH2N pathway for menaquinone biosynthesis, and this is represented by the orange histogram bars. On the other hand, Ctra and Scoe both utilize the DH6N pathway, and this is indicated by the green histogram bars. Note by relating the acronyms under the histogram bars to the pathway diagrams in Figure 6 that Ctra and Scoe utilize different minor variations of the DH6N pathway, utilizing aminodeoxyfutalosine or futalosine, respectively, as unique intermediates. Ctra exhibits a fusion of AroC and AroD, whereas Pamo has a fusion of AroC, AroD, and AroF (as indicated in red). The cumulative total of Trp residues in the chorismate pathway and the connected menaquinone pathway are given within the color-coded horizontal bars displayed at the bottom of the figure.