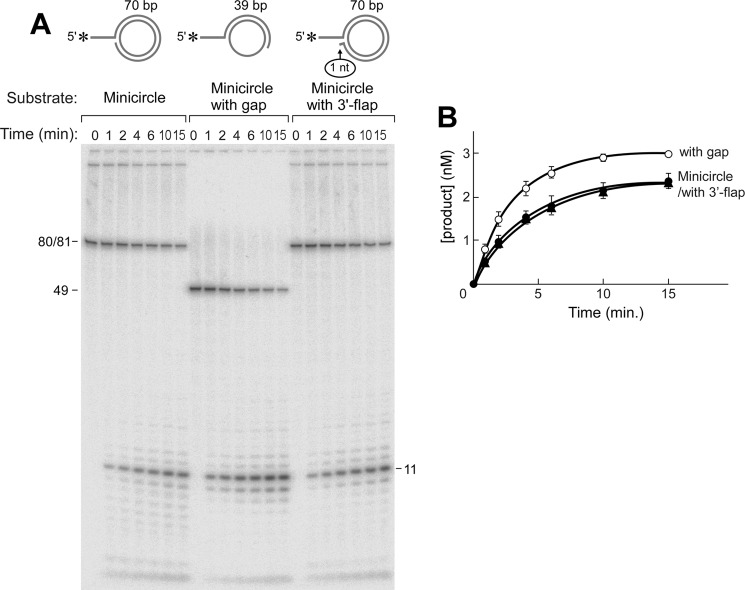
FIGURE 5.
Flap endonuclease activity of gp6 at a replication fork. Flap endonuclease assays were performed as described under “Experimental Procedures.” Reactions (10 μl) were carried out by incubating gp6 (100 nm) at 37 °C with the minicircle-based substrates (5 nm) depicted above (A). The minicircle substrate consists of an oligonucleotide having 70 nt 3′-complementary to the 70-nt minicircle and an additional portion of 10 nt 5′-non-complementary to the minicircle (A10:B5). The minicircle with gap substrate consists of an oligonucleotide having 39 nt 3′-complementary to the minicircle and 10 nt 5′-non-complementary to the minicircle (A7:B3). The minicircle with 3′-flap substrate has a 10-nt 5′-overhang and is identical to the one described in A except for an additional nucleotide at the 3′-end that results in a 1-nt 3′-flap (A12:B3). The reaction products were separated on an 18% denaturing polyacrylamide gel containing 7 m urea. Completely denatured strands are labeled on the right side of the autoradiograph. The smearing observed above those bands represents incomplete denaturation of substrates. B, the amount of oligoribonucleotide cleaved by gp6 in the assays in A was analyzed quantitatively using a Fuji BAS 1000 Bioimaging analyzer and presented in the graph. Plots represent the mean, and error bars indicate S.D. from three independent experiments.