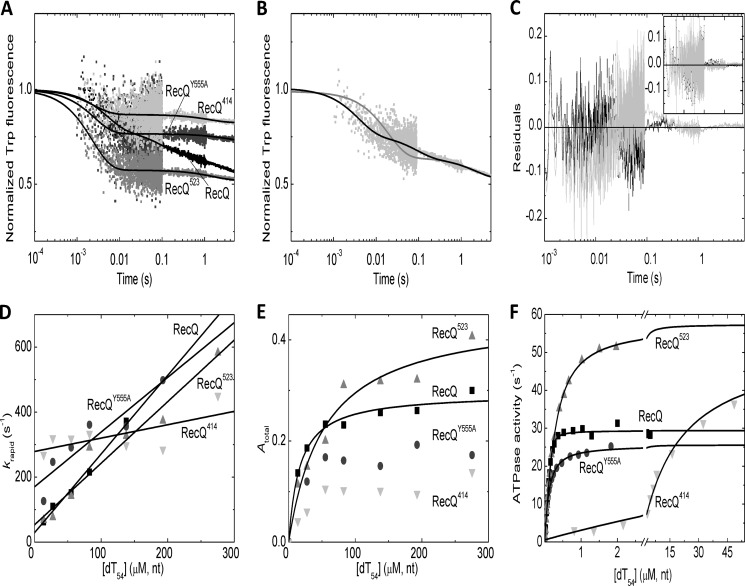
FIGURE 4.
DNA binding and DNA-activated ATPase activity of wild-type and mutant RecQ constructs. A, stopped-flow Trp fluorescence traces of the interaction of RecQ constructs (wild-type RecQ (black), RecQY555A (dark gray), RecQ523 (gray), and RecQ414 (light gray); all proteins applied at 0. 5 μm) with dT54 (81 μm, nt) at 5 °C. Fluorescence levels were normalized to that of the DNA-free state. Lines show triple (wild-type RecQ) or double exponential (all mutants) fits to the data sets. (The slowest phase arose from photobleaching in each case; see “Results.”) B and C, comparison of fits comprising one (B, gray line) and two (B, black line) exponential DNA-binding phases (in addition to the slow photobleaching phase) to the wild-type RecQ trace (cf. A). Residuals of the fits shown in the main panel of C (gray and black, respectively) indicated systematic deviation from data points when fitting a single binding phase (gray). The inset of C shows that, in the case of RecQ523, the residuals of fits comprising either one (gray) or two (black) binding phases showed no systematic deviation from data points (fitted lines were indistinguishable; cf. A). This finding applied to all mutant constructs. D and E, dT54 concentration dependence of rapid phase rate constants (krapid; D) and total amplitudes (Atotal; E) of transients recorded at 5 °C as in A (wild-type RecQ (black squares), RecQY555A (dark gray circles), RecQ523 (gray triangles), and RecQ414 (light gray inverted triangles)) alongside linear (D) and hyperbolic (E) fits. (RecQY555A and RecQ414 amplitude data could not be reasonably fitted due to noise in the low values.) F, dT54-activated ATPase activity of RecQ constructs (wild-type RecQ 15 nm, mutant constructs 10 nm; symbols as in D) as determined by a PK-LDH assay at 25 °C. Quadratic (wild-type RecQ, RecQY555A) and hyperbolic (RecQ523 and RecQ414) fits are shown as lines. Parameters derived from fits are shown in Table 3.