Figure 3.
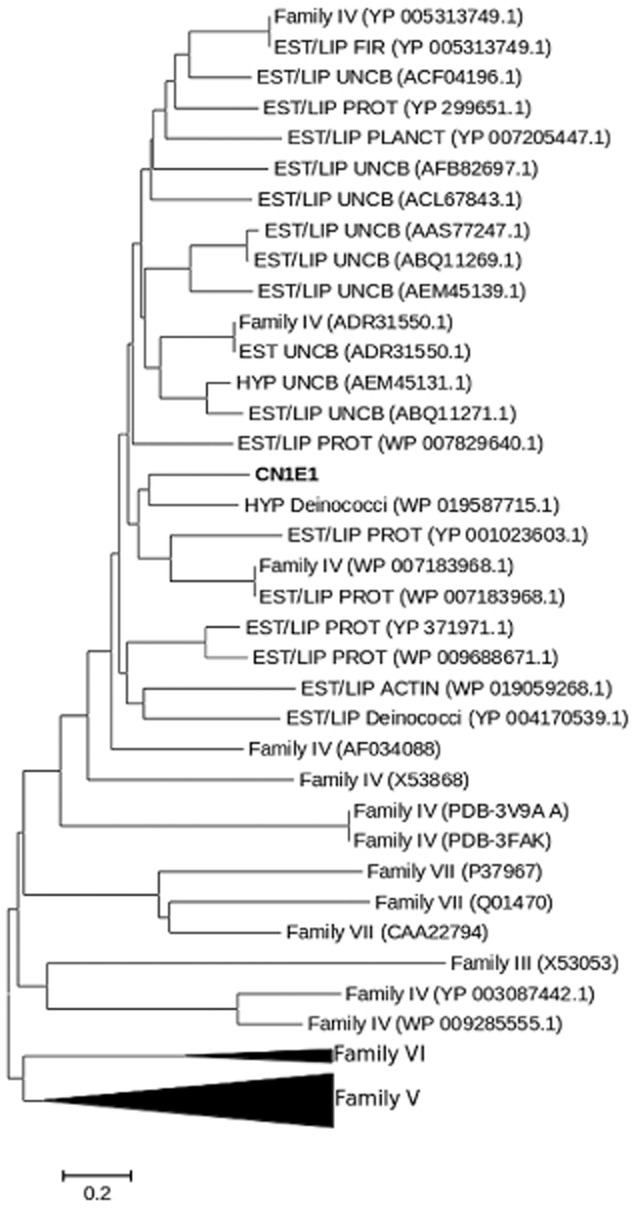
The unrooted circular neighbour-joining tree indicates the polypeptide sequence phylogenetic positions for the CN1E1 enzyme (in boldface) and reference hydrolases. The tree was constructed using an aligned 297 AA-long sequence. The GenBank and PDB accession numbers are indicated in brackets. For the dendrogram construction details, see Martínez-Martínez and colleagues (2013) and Tamura and colleagues (2007). The Family V cluster includes the sequences with the following accession numbers: YP960710.1, ADP98993.1, ZP01735705.1, ZP01735705.1, CAE54381.1, ZP01307774.1, X53869, AEO74498.1, YP001347584.1, NP251639.1, YP790224.1, YP0029804424.1, YP002909304.2, YP001810250.1, ZP03570306.1, YP299126.1, YP583166.1, YP002005156.1, 725653.1 and YP006884923.1. The Family VI cluster includes YP007541785, WP007625024, WP008294645, S78600 and PDB 3CN7. The scale bar represents 0.2 substitutions per position. The lipase/esterase families are depicted based on the Arpigny and Jaeger (1999) classification system. The abbreviations are as follows: EST, esterase; LIP, lipase; HYP, hypothetical protein; ACTIN, Actinobacteria; FIR, Firmicutes; PLANCT, Planctomycetes; PROT, Proteobacteria; UNCB, uncultured bacterium.
